6DBT
 
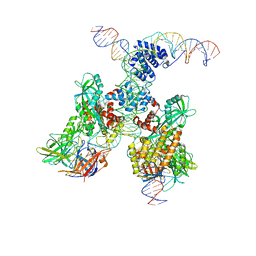 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBU
 
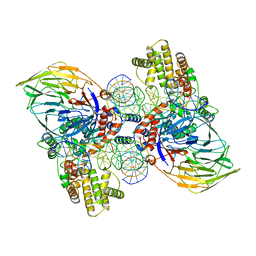 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBV
 
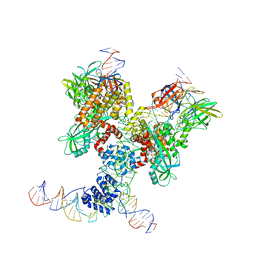 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBO
 
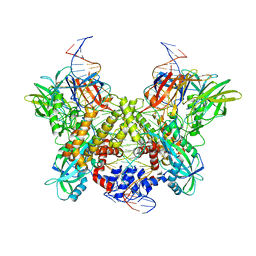 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBW
 
 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBX
 
 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7UPN
 
 | |
6CVP
 
 | | Human Aprataxin (Aptx) R199H bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVS
 
 | | Human Aprataxin (Aptx) L248M bound to DNA, AMP and Zn product | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, BETA-MERCAPTOETHANOL, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVR
 
 | | Human Aprataxin (Aptx) S242N bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVQ
 
 | | Human Aprataxin (Aptx) H201Q bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, BETA-MERCAPTOETHANOL, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVO
 
 | | Human Aprataxin (Aptx) bound to nicked RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, BETA-MERCAPTOETHANOL, ... | | Authors: | Schellenberg, M.J, Tumbale, P.P, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVT
 
 | | Human Aprataxin (Aptx) V263G bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
3U7F
 
 | | Crystal structure of mPNKP catalytic fragment (D170A) bound to single-stranded DNA (TCCTCp) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, DNA, GLYCEROL, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U7G
 
 | | Crystal structure of mPNKP catalytic fragment (D170A) bound to single-stranded DNA (TCCTAp) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, DNA, GLYCEROL, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U7H
 
 | | Crystal structure of mPNKP catalytic fragment (D170A) bound to single-stranded DNA (TCCTTp) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, DNA, GLYCEROL, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4CI1
 
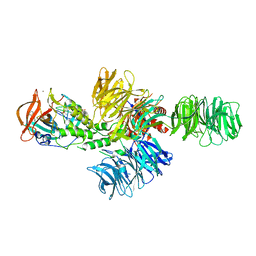 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to thalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Thalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4M6F
 
 | |
4WHV
 
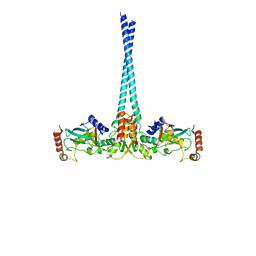 | | E3 ubiquitin-protein ligase RNF8 in complex with Ubiquitin-conjugating enzyme E2 N and Polyubiquitin-B | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-09-23 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (8.3 Å) | | Cite: | RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J.Biol.Chem., 291, 2016
|
|
7LT3
 
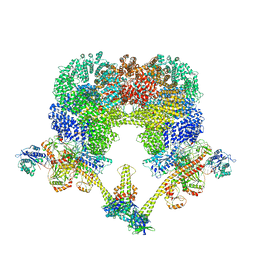 | | NHEJ Long-range synaptic complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | Authors: | He, Y, Chen, S. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of long-range to short-range synaptic transition in NHEJ.
Nature, 593, 2021
|
|
7KZV
 
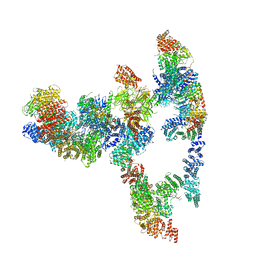 | |
7KZT
 
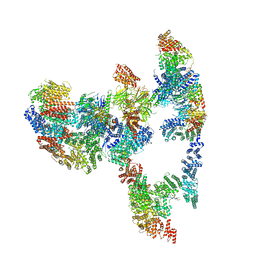 | |
7KZS
 
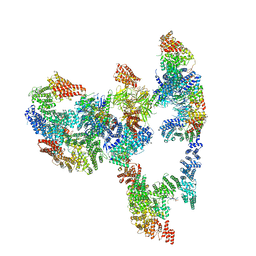 | |
2ZO2
 
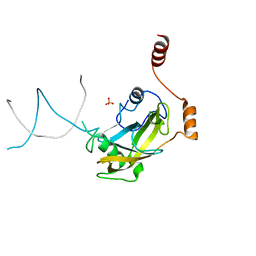 | | Mouse NP95 SRA domain non-specific DNA complex | | Descriptor: | DNA (5'-D(*DAP*DAP*DCP*DTP*DGP*DCP*DGP*DCP*DAP*DGP*DTP*DT)-3'), E3 ubiquitin-protein ligase UHRF1, PHOSPHATE ION | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
2ZO0
 
 | | mouse NP95 SRA domain DNA specific complex 1 | | Descriptor: | DNA (5'-D(*DGP*DTP*DCP*DAP*DGP*(5CM)P*DGP*DCP*DAP*DAP*DTP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DAP*DTP*DGP*DCP*DGP*DCP*DTP*DGP*DAP*DC)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
