8Q9Q
 
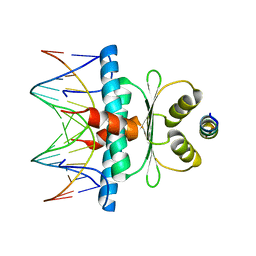 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC7 deacetylase binding motif | | Descriptor: | HDAC7 (histone deacetylase 7) binding motif peptide: GLY-VAL-VAL-LYS-GLN-LYS-LEU-ALA-GLU-VAL-ILE-LEU-LYS-LYS-GLN, MADS box dsDNA: AACTATTTATAAGA, MADS box dsDNA: TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
6ASD
 
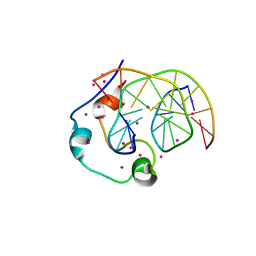 | | Zinc finger region of human TET1 in complex with CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3'), Methylcytosine dioxygenase TET1, UNKNOWN ATOM OR ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
4TPS
 
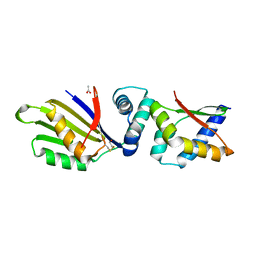 | | Sporulation Inhibitor of DNA Replication, SirA, in complex with Domain I of DnaA | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, Chromosomal replication initiator protein DnaA, ... | | Authors: | Jameson, K.H, Turkenburg, J.P, Fogg, M.J, Grahl, A, Wilkinson, A.J. | | Deposit date: | 2014-06-09 | | Release date: | 2014-07-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and interactions of the Bacillus subtilis sporulation inhibitor of DNA replication, SirA, with domain I of DnaA.
Mol.Microbiol., 93, 2014
|
|
8CMN
 
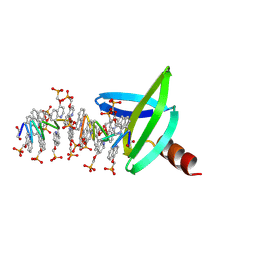 | | 18mer DNA mimic Foldamer with an aliphatic linker in complex with Sac7d wild protein | | Descriptor: | DNA-binding protein 7d, N-[2-(2-methyl-1,3-dioxolan-2-yl)phenyl]-2-{[5-(trifluoromethyl)pyridin-2-yl]amino}pyridine-4-carboxamide | | Authors: | Deepak, D, Corvaglia, V, Wu, J, Huc, I. | | Deposit date: | 2023-02-20 | | Release date: | 2023-07-19 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA Mimic Foldamer Recognition of a Chromosomal Protein.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
5V3M
 
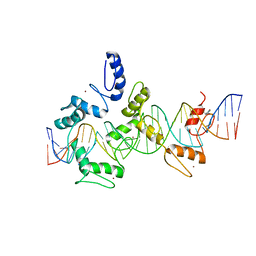 | | mouseZFP568-ZnF1-11 in complex with DNA | | Descriptor: | DNA (28-MER), ZINC ION, Zinc finger protein 568 | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
6VAE
 
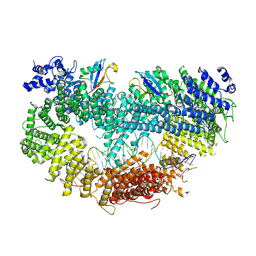 | |
4P4P
 
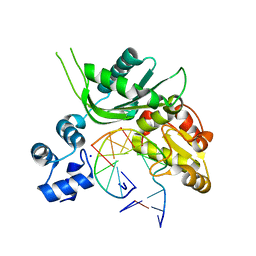 | | Crystal structure of Leishmania infantum polymerase beta: Nick complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(P*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Mejia, E, Burak, M, Alonso, A, Larraga, V, Kunkel, T, Bebenek, K, Garcia-Diaz, M. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2973 Å) | | Cite: | Structures of the Leishmania infantum polymerase beta.
DNA Repair (Amst.), 18, 2014
|
|
6ASB
 
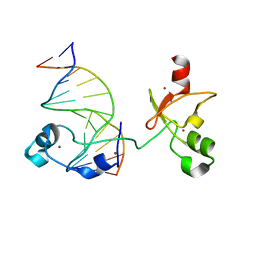 | | CXXC and PHD-type zinc finger regions of FBXL19 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*TP*TP*GP*GP*C)-3'), F-box/LRR-repeat protein 19, ZINC ION | | Authors: | Liu, K, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
2R5U
 
 | |
3ULP
 
 | | Plasmodium falciparum SSB complex with ssDNA | | Descriptor: | DNA (35-MER), Single-strand binding protein | | Authors: | Antony, E, Lohman, T.M, Korolev, S. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasmodium falciparum SSB Tetramer Wraps Single-Stranded DNA with Similar Topology but Opposite Polarity to E. coli SSB.
J.Mol.Biol., 420, 2012
|
|
9FZ6
 
 | | A 2.58A crystal structure of S. aureus DNA gyrase and DNA with metals identified through anomalous scattering | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G)-3'), DNA (5'-D(*AP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Morgan, H, Duman, R, Bax, B.D, Warren, A.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | How Do Gepotidacin and Zoliflodacin Stabilize DNA Cleavage Complexes with Bacterial Type IIA Topoisomerases? 1. Experimental Definition of Metal Binding Sites.
Int J Mol Sci, 25, 2024
|
|
7Z8S
 
 | | Mot1:TBP:DNA - post hydrolysis state | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z7N
 
 | | Mot1E1434Q:TBP:DNA - substrate recognition state | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZB5
 
 | | Mot1(1-1836):TBP:DNA - post-hydrolysis complex dimer | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4RI9
 
 | |
4RIA
 
 | | FAN1 Nuclease bound to 5' phosphorylated nicked DNA | | Descriptor: | BARIUM ION, DNA (5'-D(*TP*TP*TP*TP*TP*TP*G*AP*GP*GP*CP*GP*TP*G)-3'), DNA (5'-D(P*AP*GP*AP*CP*TP*CP*CP*TP*C)-3'), ... | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
4RIC
 
 | | FAN1 Nuclease bound to 5' hydroxyl (dT-dT) single flap DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*AP*GP*GP*AP*GP*TP*CP*T)-3'), DNA (5'-D(*TP*TP*AP*GP*CP*CP*AP*CP*GP*CP*CP*TP*AP*GP*AP*CP*TP*CP*CP*TP*C)-3'), ... | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
4RIB
 
 | | FAN1 Nuclease bound to 5' phosphorylated p(dT) single flap DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*AP*GP*GP*AP*GP*TP*CP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*TP*GP*AP*GP*GP*CP*GP*TP*G)-3'), ... | | Authors: | Pavletich, N.P, Wang, R. | | Deposit date: | 2014-10-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1.
Science, 346, 2014
|
|
4RI8
 
 | |
8CE2
 
 | | X-ray structure of the adduct formed upon reaction of a B-DNA double helical dodecamer with dirhodium tetraacetate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Tito, G, Troisi, R, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-02-01 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Dirhodium tetraacetate binding to a B-DNA double helical dodecamer probed by X-ray crystallography and mass spectrometry.
Dalton Trans, 52, 2023
|
|
2HCB
 
 | | Structure of AMPPCP-bound DnaA from Aquifex aeolicus | | Descriptor: | Chromosomal replication initiator protein dnaA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Erzberger, J.P, Mott, M.L, Berger, J.M. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling.
Nat.Struct.Mol.Biol., 13, 2006
|
|
8G0H
 
 | |
7CCJ
 
 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*AP*TP*C)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Zhao, G, Gan, J, Liu, G, Wu, G, He, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
7CCD
 
 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*GP*AS*AP*CP*GP*TP*G)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Li, J, Liu, G, Zhao, G, Wang, Y, Hu, W, Deng, Z, Gan, J, Zhao, Y, He, X. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
6EN0
 
 | |
