6SUD
 
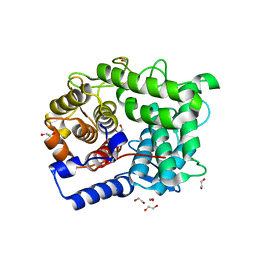 | | Structure of L320A mutant of Rex8A from Paenibacillus barcinonensis complexed with xylose. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Reducing-end xylose-releasing exo-oligoxylanase Rex8A, ... | | Authors: | Jimenez-Ortega, E, Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-09-13 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural analysis of the reducing-end xylose-releasing exo-oligoxylanase Rex8A from Paenibacillus barcinonensis BP-23 deciphers its molecular specificity.
Febs J., 287, 2020
|
|
6SRD
 
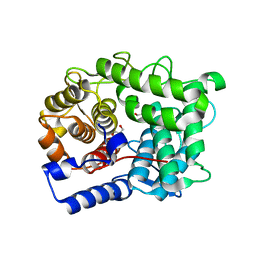 | |
7DFS
 
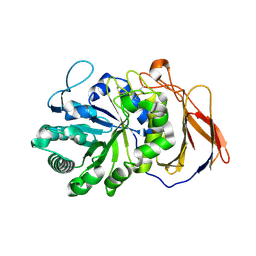 | | Crystal structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S - Rha-GlcA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase, alpha-D-mannopyranose, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
1GCY
 
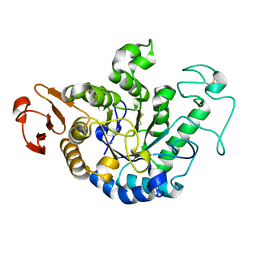 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF MALTOTETRAOSE-FORMING EXO-AMYLASE | | Descriptor: | CALCIUM ION, GLUCAN 1,4-ALPHA-MALTOTETRAHYDROLASE | | Authors: | Mezaki, Y, Katsuya, Y, Kubota, M, Matsuura, Y. | | Deposit date: | 2000-08-14 | | Release date: | 2000-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization and structural analysis of intact maltotetraose-forming exo-amylase from Pseudomonas stutzeri.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
8PXI
 
 | | Crystal structure of Endothiapepsin soaked with FRG283 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-{[methyl(prop-2-yn-1-yl)amino]methyl}-1,3-thiazol-4-yl)piperidin-4-ol, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-23 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
3C1U
 
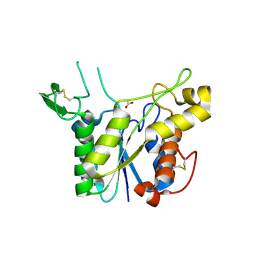 | | D192N mutant of Rhamnogalacturonan acetylesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Rhamnogalacturonan acetylesterase | | Authors: | Langkilde, A, Lo Leggio, L, Navarro Poulsen, J.C, Molgaard, A, Larsen, S. | | Deposit date: | 2008-01-24 | | Release date: | 2008-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Short strong hydrogen bonds in proteins: a case study of rhamnogalacturonan acetylesterase
ACTA CRYSTALLOGR.,SECT.D, 64, 2008
|
|
5E4J
 
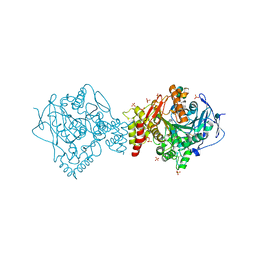 | | Acetylcholinesterase Methylene Blue no PEG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, DECAMETHONIUM ION, ... | | Authors: | Dym, O. | | Deposit date: | 2015-10-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The impact of crystallization conditions on structure-based drug design: A case study on the methylene blue/acetylcholinesterase complex.
Protein Sci., 25, 2016
|
|
8EJX
 
 | |
6T9A
 
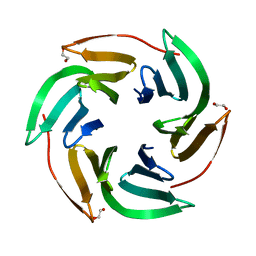 | | Crystal structrue of RSL W31FW76F lectin mutant in complex with L-fucose | | Descriptor: | 1,2-ETHANEDIOL, Fucose-binding lectin protein, alpha-L-fucopyranose, ... | | Authors: | Houser, J, Kozmon, S, Wimmerova, M. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The CH-pi Interaction in Protein-Carbohydrate Binding: Bioinformatics and In Vitro Quantification.
Chemistry, 26, 2020
|
|
6T99
 
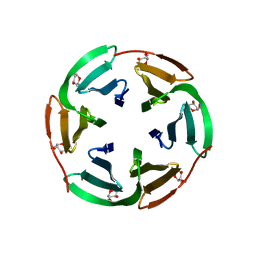 | |
8OT2
 
 | | Structural and functional studies of geldanamycin amide synthase ShGdmF | | Descriptor: | 1,2-ETHANEDIOL, 3-azanyl-5-methyl-phenol, ACETATE ION, ... | | Authors: | Ewert, W, Zeilinger, C, Kirschning, A, Preller, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional studies of geldanamycin amide synthase ShGdmF
To Be Published
|
|
6T9B
 
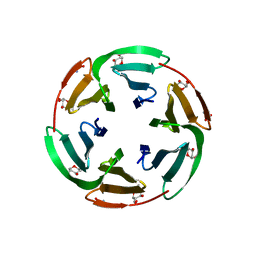 | | Crystal structrue of RSL W31A lectin mutant in complex with alpha-methylfucoside | | Descriptor: | Fucose-binding lectin protein, GLYCINE, methyl alpha-L-fucopyranoside | | Authors: | Houser, J, Komarek, J, Kozmon, S, Wimmerova, M. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The CH-pi Interaction in Protein-Carbohydrate Binding: Bioinformatics and In Vitro Quantification.
Chemistry, 26, 2020
|
|
8PKC
 
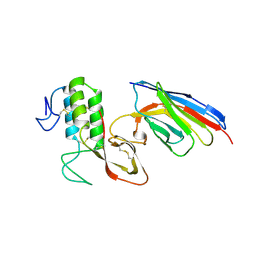 | |
8GW5
 
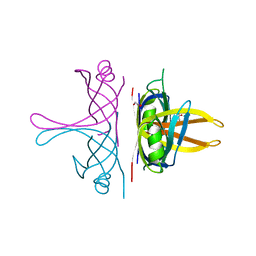 | |
1O7A
 
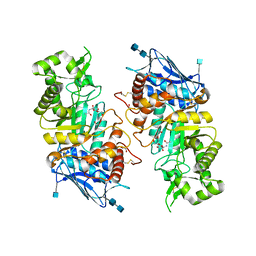 | | Human beta-Hexosaminidase B | | Descriptor: | 1,2-ETHANEDIOL, 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maier, T, Strater, N, Schuette, C, Klingenstein, R, Sandhoff, K, Saenger, W. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The X-Ray Crystal Structure of Human Beta-Hexosaminidase B Provides New Insights Into Sandhoff Disease
J.Mol.Biol., 328, 2003
|
|
3C9L
 
 | |
5E2I
 
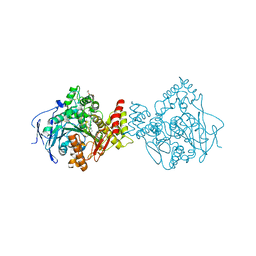 | | Acetylcholinesterase Methylene Blue no PEG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, DECAMETHONIUM ION, ... | | Authors: | Dym, O. | | Deposit date: | 2015-10-01 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The impact of crystallization conditions on structure-based drug design: A case study on the methylene blue/acetylcholinesterase complex.
Protein Sci., 25, 2016
|
|
6HIL
 
 | |
1IZ2
 
 | | Interactions causing the kinetic trap in serpin protein folding | | Descriptor: | alpha-D-glucopyranose-(1-2)-(5R)-5-[(2R)-2-hydroxynonyl]-beta-D-xylulofuranose, alpha1-antitrypsin | | Authors: | Im, H, Woo, M.-S, Hwang, K.Y, Yu, M.-H. | | Deposit date: | 2002-09-19 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions causing the kinetic trap in serpin protein folding
J.BIOL.CHEM., 277, 2002
|
|
8GW0
 
 | | Crystal structure of the human dihydroorotase domain in complex with malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CAD protein, ZINC ION | | Authors: | Yang, P.C, Liu, H.W, Huang, H.Y, Huang, C.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Complexed Crystal Structure of the Dihydroorotase Domain of Human CAD Protein with the Anticancer Drug 5-Fluorouracil.
Biomolecules, 13, 2023
|
|
8GVZ
 
 | | Crystal structure of the human dihydroorotase domain in complex with the anticancer drug 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, CAD protein, ZINC ION | | Authors: | Liu, H.W, Yang, P.C, Huang, H.Y, Huang, C.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Complexed Crystal Structure of the Dihydroorotase Domain of Human CAD Protein with the Anticancer Drug 5-Fluorouracil.
Biomolecules, 13, 2023
|
|
6HX7
 
 | | Crystal structure of human R180T variant of ORNITHINE AMINOTRANSFERASE at 1.8 Angstrom | | Descriptor: | Ornithine aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Giardina, G, Montioli, R, Cellini, B, Cutruzzola, F, Borri Voltattorni, C. | | Deposit date: | 2018-10-16 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | R180T variant of delta-ornithine aminotransferase associated with gyrate atrophy: biochemical, computational, X-ray and NMR studies provide insight into its catalytic features.
Febs J., 286, 2019
|
|
6HIK
 
 | |
5DWU
 
 | | Beta common receptor in complex with a Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, Fab - Heavy Chain, ... | | Authors: | Dhagat, U, Parker, M.W. | | Deposit date: | 2015-09-23 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | CSL311, a novel, potent, therapeutic monoclonal antibody for the treatment of diseases mediated by the common beta chain of the IL-3, GM-CSF and IL-5 receptors.
Mabs, 8, 2016
|
|
6MV1
 
 | | 2.15A resolution structure of the CS-b5R domains of human Ncb5or (NAD+ form) | | Descriptor: | Cytochrome b5 reductase 4, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Benson, D.R, Cooper, A, Gao, P, Zhu, H. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of the naturally fused CS and cytochrome b5reductase (b5R) domains of Ncb5or reveal an expanded CS fold, extensive CS-b5R interactions and productive binding of the NAD(P)+nicotinamide ring.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
