3AL0
 
 | | Crystal structure of the glutamine transamidosome from Thermotoga maritima in the glutamylation state. | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Glutamyl-tRNA(Gln) amidotransferase subunit A, Glutamyl-tRNA(Gln) amidotransferase subunit C,Linker,Glutamate--tRNA ligase 2, ... | | Authors: | Ito, T, Yokoyama, S. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.368 Å) | | Cite: | Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions.
Nature, 467, 2010
|
|
3G5H
 
 | | Crystallographic analysis of cytochrome P450 cyp121 | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Belin, P, Le Du, M.H, Gondry, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1A7T
 
 | | METALLO-BETA-LACTAMASE WITH MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, METALLO-BETA-LACTAMASE, SODIUM ION, ... | | Authors: | Fitzgerald, P.M.D, Wu, J.K, Toney, J.H. | | Deposit date: | 1998-03-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unanticipated inhibition of the metallo-beta-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-A resolution.
Biochemistry, 37, 1998
|
|
1ON3
 
 | | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with methylmalonyl-coenzyme a and methylmalonic acid bound) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CADMIUM ION, METHYLMALONIC ACID, ... | | Authors: | Hall, P.R, Wang, Y.-F, Rivera-Hainaj, R.E, Zheng, X, Pustai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core
Embo J., 22, 2003
|
|
1W7N
 
 | | Crystal structure of human kynurenine aminotransferase I in PMP form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, KYNURENINE--OXOGLUTARATE TRANSAMINASE I | | Authors: | Rossi, F, Han, Q, Li, J, Li, J, Rizzi, M. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Kynurenine Aminotransferase I
J.Biol.Chem., 279, 2004
|
|
1W4U
 
 | | NMR solution structure of the ubiquitin conjugating enzyme UbcH5B | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-17 KDA 2 | | Authors: | Houben, K, Dominguez, C, Van Schaik, F.M.A, Timmers, H.T.M, Bonvin, A.M.J.J, Boelens, R. | | Deposit date: | 2004-07-29 | | Release date: | 2004-11-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ubiquitin-Conjugating Enzyme Ubch5B
J.Mol.Biol., 344, 2004
|
|
1W7M
 
 | | Crystal structure of human kynurenine aminotransferase I in complex with L-Phe | | Descriptor: | KYNURENINE--OXOGLUTARATE TRANSAMINASE I, PHENYLALANINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Han, Q, Li, J, Li, J, Rizzi, M. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Kynurenine Aminotransferase I
J.Biol.Chem., 279, 2004
|
|
2ALG
 
 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, HEXAETHYLENE GLYCOL, LAURIC ACID, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, J, Zanotti, G. | | Deposit date: | 2005-08-05 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
1W0G
 
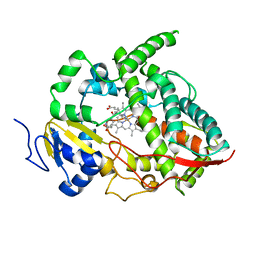 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, METYRAPONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
2HLE
 
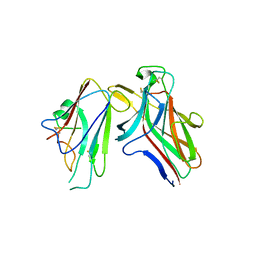 | |
1CE2
 
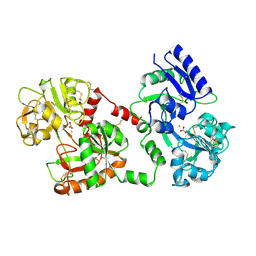 | | STRUCTURE OF DIFERRIC BUFFALO LACTOFERRIN AT 2.5A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, PROTEIN (LACTOFERRIN) | | Authors: | Karthikeyan, S, Paramasivam, M, Yadav, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-03-13 | | Release date: | 1999-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of buffalo lactoferrin at 2.5 A resolution using crystals grown at 303 K shows different orientations of the N and C lobes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2AGC
 
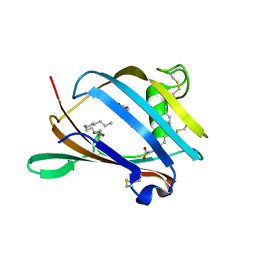 | | Crystal Structure of mouse GM2- activator Protein | | Descriptor: | Ganglioside GM2 activator, LAURIC ACID, MYRISTIC ACID | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2PJL
 
 | |
2PXR
 
 | | Crystal Structure of HIV-1 CA146 in the Presence of CAP-1 | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), ZINC ION | | Authors: | Kelly, B.N. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein.
J.Mol.Biol., 373, 2007
|
|
3RLL
 
 | | Crystal structure of the T877A androgen receptor ligand binding domain in complex with (S)-N-(4-Cyano-3-(trifluoromethyl)phenyl)-3-(4-cyanonaphthalen-1-yloxy)-2-hydroxy-2-methylpropanamide | | Descriptor: | (2S)-3-[(4-cyanonaphthalen-1-yl)oxy]-N-[4-cyano-3-(trifluoromethyl)phenyl]-2-hydroxy-2-methylpropanamide, Androgen receptor | | Authors: | Bohl, C.E, Duke, C.B, Jones, A, Dalton, J.T, Miller, D.D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected binding orientation of bulky-B-ring anti-androgens and implications for future drug targets.
J.Med.Chem., 54, 2011
|
|
2Q8S
 
 | |
2D4R
 
 | | Crystal structure of TTHA0849 from Thermus thermophilus HB8 | | Descriptor: | SULFATE ION, hypothetical protein TTHA0849 | | Authors: | Nakabayashi, M, Shibata, N, Kuramitsu, S, Higuchi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a conserved hypothetical protein, TTHA0849 from Thermus thermophilus HB8, at 2.4 A resolution: a putative member of the StAR-related lipid-transfer (START) domain superfamily.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2FOF
 
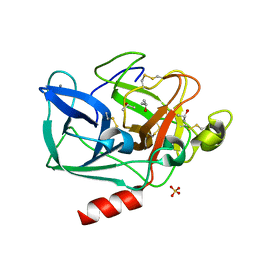 | | Structure of porcine pancreatic elastase in 80% isopropanol | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
3HBV
 
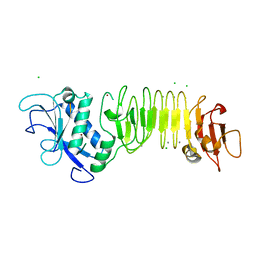 | | PrtC methionine mutants: M226A in-house | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-05 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
2Y6B
 
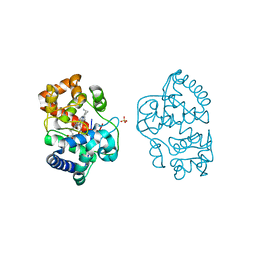 | | Ascorbate Peroxidase R38K mutant | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Metcalfe, C.L, Efimov, I, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-01-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proton Delivery to Ferryl Heme in a Heme Peroxidase: Enzymatic Use of the Grotthuss Mechanism.
J.Am.Chem.Soc., 133, 2011
|
|
2LL3
 
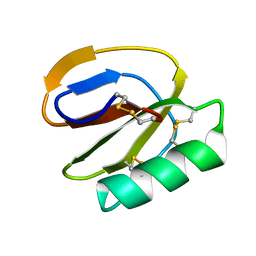 | |
3KDH
 
 | | Structure of ligand-free PYL2 | | Descriptor: | Putative uncharacterized protein At2g26040 | | Authors: | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
5NKN
 
 | | Crystal structure of an Anticalin-colchicine complex | | Descriptor: | N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide, Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A, Barkovskiy, M. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An engineered lipocalin that tightly complexes the plant poison colchicine for use as antidote and in bioanalytical applications.
Biol. Chem., 400, 2019
|
|
8P7D
 
 | | CryoEM structure of METTL6 tRNA SerRS complex in a 1:1:2 stoichiometry | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, Serine tRNA, ... | | Authors: | Throll, P, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5L05
 
 | |
