6T6W
 
 | |
6T6R
 
 | | Human endoplasmic reticulum aminopeptidase 1 (ERAP1) in complex with (4aR,5S,6R,8S,8aR)-5-(2-(Furan-3-yl)ethyl)-8-hydroxy-5,6,8a-trimethyl-3,4,4a,5,6,7,8,8a-octahydronaphthalene-1-carboxylic acid | | Descriptor: | (4~{a}~{R},5~{S},6~{R},8~{S},8~{a}~{R})-5-[2-(furan-3-yl)ethyl]-5,6,8~{a}-trimethyl-8-oxidanyl-3,4,4~{a},6,7,8-hexahydronaphthalene-1-carboxylic acid, 1,2-ETHANEDIOL, D-MALATE, ... | | Authors: | Rowland, P. | | Deposit date: | 2019-10-18 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Targeting the Regulatory Site of ER Aminopeptidase 1 Leads to the Discovery of a Natural Product Modulator of Antigen Presentation.
J.Med.Chem., 63, 2020
|
|
7E9K
 
 | | Crystal Structure of POMGNT2 in complex with UDP and mono-mannosyl peptide (379Man long peptide) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2021-03-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Genes Cells, 26, 2021
|
|
6SUV
 
 | |
6SJC
 
 | | Structure of T. thermophilus AspRS in Complex with 5'-O-(N-(L-aspartyl)-sulfamoyl)adenosine | | Descriptor: | 5'-O-(L-alpha-aspartylsulfamoyl)adenosine, Aspartate--tRNA(Asp) ligase | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
6SLN
 
 | | Structure of the RagAB peptide transporter | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Madej, M, Ranson, N.A, White, J.B.R. | | Deposit date: | 2019-08-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis.
Nat Microbiol, 5, 2020
|
|
6SMZ
 
 | |
7EFQ
 
 | | Crystal structure of hPPARgamma ligand binding domain complexed with rosiglitazone-based fluorescence probe | | Descriptor: | (5S)-5-[[4-[2-[[7-(diethylamino)-2-oxidanylidene-chromen-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Yoshikawa, C, Ishida, H, Ohashi, N, Itoh, T. | | Deposit date: | 2021-03-23 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis of a Coumarin-Based PPAR gamma Fluorescence Probe for Competitive Binding Assay.
Int J Mol Sci, 22, 2021
|
|
7ENU
 
 | | Crystal structure of iron-saturated C-terminal half of lactoferrin produced proteolytically using pepsin at 2.32A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, J, Maurya, A, Viswanathan, V, Singh, P.K, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Crystal structure of iron-saturated C-terminal half of lactoferrin produced proteolytically using pepsin at 2.32A resolution
To Be Published
|
|
6SKT
 
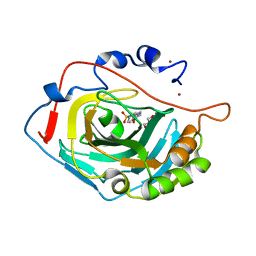 | |
7E8M
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with mutated RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ge, J.W, Wang, R.K, Lan, J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species.
Immunity, 54, 2021
|
|
7EEH
 
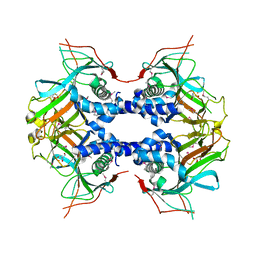 | | Selenomethionine labeled Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TqaL | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, TqaL | | Authors: | Bunno, R, Mori, T, Awakawa, T, Abe, I. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-26 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aziridine Formation by a Fe II / alpha-Ketoglutarate Dependent Oxygenase and 2-Aminoisobutyrate Biosynthesis in Fungi.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6SLW
 
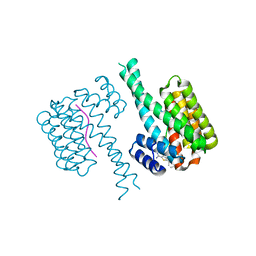 | | Fragment AZ-004 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-methyl-5-phenyl-thiophene-2-carboximidamide, WW domain-containing transcription regulator protein 1 | | Authors: | Ottmann, C, Wolter, M, Guillory, X, Leysen, S, Genet, S, Somsen, B, Patel, J, Castaldi, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6SXT
 
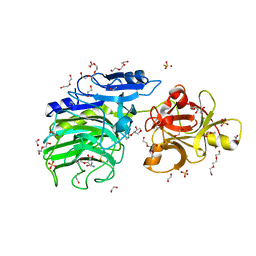 | | GH54 a-l-arabinofuranosidase soaked with aziridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGregor, N.G.S, Davies, G.J, Nin-Hill, A, Rovira, C. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | Rational Design of Mechanism-Based Inhibitors and Activity-Based Probes for the Identification of Retaining alpha-l-Arabinofuranosidases.
J.Am.Chem.Soc., 142, 2020
|
|
6SM7
 
 | | Crystal structure of SLA Reductase YihU from E. Coli | | Descriptor: | 3-sulfolactaldehyde reductase, BORIC ACID | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dynamic Structural Changes Accompany the Production of Dihydroxypropanesulfonate by Sulfolactaldehyde Reductase
Acs Catalysis, 2020
|
|
6SNQ
 
 | | Crystal structures of human PGM1 isoform 2 | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Phosphoglucomutase-1, ZINC ION | | Authors: | Backe, P.H, Laerdahl, J.K, Kittelsen, L.S, Dalhus, B, Morkrid, L, Bjoras, M. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for substrate and product recognition in human phosphoglucomutase-1 (PGM1) isoform 2, a member of the alpha-D-phosphohexomutase superfamily.
Sci Rep, 10, 2020
|
|
7DWS
 
 | |
6SPI
 
 | | A4V MUTANT OF HUMAN SOD1 WITH EBSELEN DERIVATIVE 4 | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION, ... | | Authors: | Chantadul, V, Amporndanai, K, Wright, G, Antonyuk, S, Hasnain, S. | | Deposit date: | 2019-09-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ebselen as template for stabilization of A4V mutant dimer for motor neuron disease therapy.
Commun Biol, 3, 2020
|
|
6SSQ
 
 | | Crystal structure of RARbeta LBD in complex with LG 100754 | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, CITRATE ANION, GLYCEROL, ... | | Authors: | le Maire, A, Teyssier, C, Germain, P, Bourguet, W. | | Deposit date: | 2019-09-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of RXR-RAR Heterodimers by RXR- and RAR-Specific Ligands and Their Combinations.
Cells, 8, 2019
|
|
6ST0
 
 | |
6STJ
 
 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Cystatin domain-containing protein, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
6SQ9
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 3-hydroxynaphthalene-2-carboxylic acid | | Descriptor: | 3-hydroxynaphthalene-2-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6SQN
 
 | |
6SXV
 
 | | GH51 a-l-arabinofuranosidase soaked with aziridine inhibitor | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GH51 a-l-arabinofuranosidase, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Rational Design of Mechanism-Based Inhibitors and Activity-Based Probes for the Identification of Retaining alpha-l-Arabinofuranosidases.
J.Am.Chem.Soc., 142, 2020
|
|
7CWP
 
 | |
