1L9P
 
 | |
1YVR
 
 | | Ro autoantigen | | Descriptor: | 60-kDa SS-A/Ro ribonucleoprotein | | Authors: | Stein, A.J, Fuchs, G, Fu, C, Wolin, S.L, Reinisch, K.M. | | Deposit date: | 2005-02-16 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into RNA Quality Control: The Ro Autoantigen Binds Misfolded RNAs via Its Central Cavity
Cell(Cambridge,Mass.), 121, 2005
|
|
1PY9
 
 | | The crystal structure of an autoantigen in multiple sclerosis | | Descriptor: | Myelin-oligodendrocyte glycoprotein, SULFATE ION | | Authors: | Clements, C.S, Reid, H.H, Beddoe, T, Tynan, F.E, Perugini, M.A, Johns, T.G, Bernard, C.C, Rossjohn, J. | | Deposit date: | 2003-07-08 | | Release date: | 2003-09-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of myelin oligodendrocyte glycoprotein, a key autoantigen in multiple sclerosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1PZ1
 
 | |
1PYF
 
 | |
1RHH
 
 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
1JPU
 
 | | Crystal Structure of Bacillus Stearothermophilus Glycerol Dehydrogenase | | Descriptor: | ZINC ION, glycerol dehydrogenase | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
1P4E
 
 | | Flpe W330F mutant-DNA Holliday Junction Complex | | Descriptor: | 33-MER, 5'-D(*TP*AP*AP*GP*TP*TP*CP*CP*TP*AP*TP*TP*C)-3', 5'-D(*TP*TP*TP*AP*AP*AP*AP*GP*AP*AP*TP*AP*GP*GP*AP*AP*CP*TP*TP*C)-3', ... | | Authors: | Rice, P.A, Chen, Y. | | Deposit date: | 2003-04-23 | | Release date: | 2003-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the conserved Trp330 in Flp-mediated recombination. Functional and structural analysis
J.Biol.Chem., 278, 2003
|
|
1P6R
 
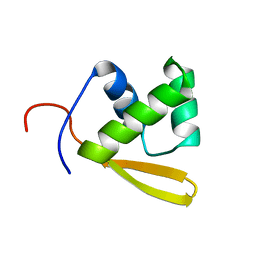 | | Solution structure of the DNA binding domain of the repressor BlaI. | | Descriptor: | Penicillinase repressor | | Authors: | Melckebeke, H.V, Vreuls, C, Gans, P, Llabres, G, Filee, P, Joris, B, Simorre, J.P. | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structural study of BlaI: implications for the repression of genes involved in beta-lactam antibiotic resistance.
J.Mol.Biol., 333, 2003
|
|
1JQ5
 
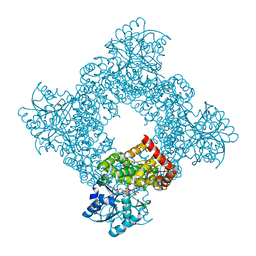 | | Bacillus Stearothermophilus Glycerol dehydrogenase complex with NAD+ | | Descriptor: | Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
2AL0
 
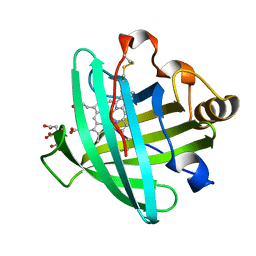 | | Crystal structure of nitrophorin 2 ferrous aqua complex | | Descriptor: | CITRIC ACID, Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-08-04 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
1ORP
 
 | |
1OYG
 
 | |
1P94
 
 | | NMR Structure of ParG symmetric dimer | | Descriptor: | plasmid partition protein ParG | | Authors: | Golovanov, A.P, Barilla, D, Golovanova, M, Hayes, F, Lian, L.Y. | | Deposit date: | 2003-05-09 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure.
Mol.Microbiol., 50, 2003
|
|
1P59
 
 | | Structure of a non-covalent Endonuclease III-DNA Complex | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*GP*(5IU)P*GP*GP*AP*C)-3', 5'-D(TP*GP*(5IU)P*CP*CP*AP*(3DR)P*GP*(5IU)P*CP*T)-3', Endonuclease III, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-04-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Trapped Endonuclease III-DNA Covalent Intermediate
Embo J., 22, 2003
|
|
1JRL
 
 | | Crystal structure of E. coli Lysophospholiase L1/Acyl-CoA Thioesterase I/Protease I L109P mutant | | Descriptor: | Acyl-CoA Thioesterase I, IMIDAZOLE, SULFATE ION | | Authors: | Lo, Y.-C, Lin, S.-C, Shaw, J.-F, Liaw, Y.-C. | | Deposit date: | 2001-08-14 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Escherichia coli Thioesterase I/Protease I/Lysophospholipase L1: Consensus Sequence Blocks Constitute the Catalytic Center of SGNH-hydrolases through a Conserved Hydrogen Bond Network
J.Mol.Biol., 330, 2003
|
|
1HPC
 
 | | REFINED STRUCTURES AT 2 ANGSTROMS AND 2.2 ANGSTROMS OF THE TWO FORMS OF THE H-PROTEIN, A LIPOAMIDE-CONTAINING PROTEIN OF THE GLYCINE DECARBOXYLASE | | Descriptor: | 5-[(3S)-1,2-dithiolan-3-yl]pentanoic acid, H PROTEIN OF THE GLYCINE CLEAVAGE SYSTEM, LIPOIC ACID | | Authors: | Pares, S, Cohen-Addad, C, Sieker, L, Neuburger, M, Douce, R. | | Deposit date: | 1994-02-17 | | Release date: | 1995-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures at 2 and 2.2 A resolution of two forms of the H-protein, a lipoamide-containing protein of the glycine decarboxylase complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1RCF
 
 | | STRUCTURE OF THE TRIGONAL FORM OF RECOMBINANT OXIDIZED FLAVODOXIN FROM ANABAENA 7120 AT 1.40 ANGSTROMS RESOLUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Burkhart, B, Ramakrishnan, B, Yan, H, Reedstrom, R, Markley, J, Straus, N, Sundaralingam, M. | | Deposit date: | 1994-10-31 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the trigonal form of recombinant oxidized flavodoxin from Anabaena 7120 at 1.40 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1JJE
 
 | |
1ZY0
 
 | | X-ray structure of peptide deformylase from Arabidopsis thaliana (AtPDF1A); crystals grown in PEG-6000 | | Descriptor: | Peptide deformylase, mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Juillan-Binard, C, Serero, A, Dardel, F, Giglione, C, Meinnel, T, Ferrer, J.-L. | | Deposit date: | 2005-06-09 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of mitochondrial (Type 1A) peptide deformylase provides clear guidelines for the design of inhibitors specific for the bacterial forms
J.Biol.Chem., 280, 2005
|
|
1IVC
 
 | | STRUCTURES OF AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(ACETYLAMINO)-5-AMINO-3-HYDROXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of aromatic inhibitors of influenza virus neuraminidase.
Biochemistry, 34, 1995
|
|
2A63
 
 | |
1QJH
 
 | |
1QNN
 
 | |
2ASN
 
 | | Crystal structure of D1A mutant of nitrophorin 2 complexed with imidazole | | Descriptor: | IMIDAZOLE, Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-08-23 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
