7Z6L
 
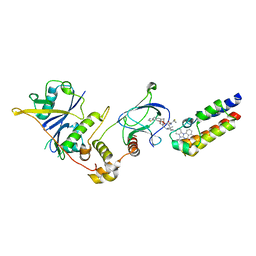 | | Crystal structure of PROTAC 5 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[3-[4-(4-bromanyl-7-cyclopentyl-5-oxidanylidene-benzimidazolo[1,2-a]quinazolin-9-yl)piperidin-1-yl]propoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Roy, M.J, Bader, G, Farnaby, W, Ciulli, A. | | Deposit date: | 2022-03-12 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
8EFG
 
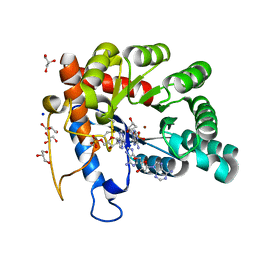 | | Crystal structure of human TATDN1 bound to dAMP and two zinc ions | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ADENINE, ... | | Authors: | Dorival, J, Eichman, B.F. | | Deposit date: | 2022-09-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human and bacterial TatD enzymes exhibit apurinic/apyrimidinic (AP) endonuclease activity.
Nucleic Acids Res., 51, 2023
|
|
8ETL
 
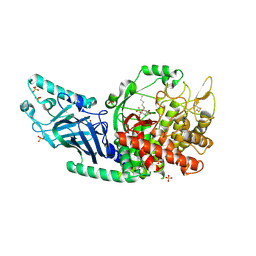 | | Co-crystal structure of Chaetomium glucosidase with compound 24 | | Descriptor: | (1S,2S,3R,4S,5S)-5-(butylamino)-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
1E0O
 
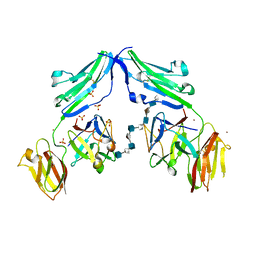 | | CRYSTAL STRUCTURE OF A TERNARY FGF1-FGFR2-HEPARIN COMPLEX | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, FIBROBLAST GROWTH FACTOR 1, FIBROBLAST GROWTH FACTOR RECEPTOR 2, ... | | Authors: | Pellegrini, L, Burke, D.F, von Delft, F, Mulloy, B, Blundell, T.L. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Fibroblast Growth Factor Receptor Ectodomain Bound to Ligand and Heparin
Nature, 407, 2000
|
|
7N64
 
 | | SARS-CoV-2 Spike (2P) in complex with G32R7 Fab (RBD and NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G32R7 Fab heavy chain, ... | | Authors: | Windsor, I.W, Jenni, S, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
1E4O
 
 | | Phosphorylase recognition and phosphorolysis of its oligosaccharide substrate: answers to a long outstanding question | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Watson, K.A, McCleverty, C, Geremia, S, Cottaz, S, Driguez, H, Johnson, L.N. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phosphorylase Recognition and Phosphorylysis of its Oligosaccharide Substrate: Answers to a Long Outstanding Question
Embo J., 18, 1999
|
|
8HPM
 
 | | LpqY-SugABC in state 2 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPN
 
 | | LpqY-SugABC in state 3 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPS
 
 | | LpqY-SugABC in state 5 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8EUR
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 26 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethyl (2-{[(1S,2S,3R,4S,5S)-2,3,4,5-tetrahydroxy-5-(hydroxymethyl)cyclohexyl]amino}ethyl)carbamate, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
7PUW
 
 | | Crystal structure of carbonic anhydrase XII with methyl 2-chloro-4-[(2-phenylethyl)sulfanyl]-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, ZINC ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
4TKN
 
 | | Structure of the SNX17 FERM domain bound to the second NPxF motif of KRIT1 | | Descriptor: | Krev interaction trapped protein 1, Sorting nexin-17 | | Authors: | Stiegler, A.L, Zhang, R, Liu, W, Boggon, T.J. | | Deposit date: | 2014-05-27 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Determinants for Binding of Sorting Nexin 17 (SNX17) to the Cytoplasmic Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 289, 2014
|
|
8EQQ
 
 | | Crystal structure of E.coli DsbA mutant E37A | | Descriptor: | CITRATE ANION, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
6X96
 
 | |
6X98
 
 | |
6JGO
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
7PUV
 
 | | Crystal structure of carbonic anhydrase XII with methyl 2-(benzenesulfonyl)-4-chloro-5-sulfamoylbenzoate | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 12, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Methyl 2-Halo-4-Substituted-5-Sulfamoyl-Benzoates as High Affinity and Selective Inhibitors of Carbonic Anhydrase IX.
Int J Mol Sci, 23, 2021
|
|
6X97
 
 | |
8EOC
 
 | | Crystal structure of E.coli DsbA mutant E24A/K58A | | Descriptor: | COPPER (II) ION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
7N0U
 
 | | Complex of recombinant Bet v 1 with Fab fragment of REGN5713 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, REGN5713 Fab fragment heavy chain, REGN5713 Fab fragment light chain, ... | | Authors: | Franklin, M.C, Romero Hernandez, A. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Targeting immunodominant Bet v 1 epitopes with monoclonal antibodies prevents the birch allergic response.
J.Allergy Clin.Immunol., 149, 2022
|
|
6FJE
 
 | |
6JO8
 
 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
4TOI
 
 | | Crystal structure of E.coli ribosomal protein S2 in complex with N-terminal domain of S1 | | Descriptor: | 30S ribosomal protein S2,Ribosomal protein S1, ZINC ION | | Authors: | Grishkovskaya, I, Byrgazov, K, Moll, I, Djinovic-Carugo, K. | | Deposit date: | 2014-06-05 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the interaction of protein S1 with the Escherichia coli ribosome.
Nucleic Acids Res., 43, 2015
|
|
8EIL
 
 | | C-Terminal Domain of BrxL from Acinetobacter BREX type I phage restriction system | | Descriptor: | MALONIC ACID, Protease Lon-related BREX system protein BrxL, SUCCINIC ACID | | Authors: | Doyle, L.A, Stoddard, B.L, Kaiser, B. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure, substrate binding and activity of a unique AAA+ protein: the BrxL phage restriction factor.
Nucleic Acids Res., 51, 2023
|
|
8HFY
 
 | | SARS-CoV-2 Omicron BA.1 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
