4O9G
 
 | | Crystal structure of the H51N mutant of the 3,4-ketoisomerase QdtA from Thermoanaerobacterium thermosaccharolyticum in complex with TDP-4-keto-6-deoxyglucose | | Descriptor: | (2S)-1-[3-[(2S)-2-oxidanylpropoxy]-2-[[(2S)-2-oxidanylpropoxy]methyl]-2-[[(2R)-2-oxidanylpropoxy]methyl]propoxy]propan-2-ol, 1,2-ETHANEDIOL, QdtA, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-01-02 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular architecture of QdtA, a sugar 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum.
Protein Sci., 23, 2014
|
|
1MRJ
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
2GYI
 
 | | DESIGN, SYNTHESIS, AND CHARACTERIZATION OF A POTENT XYLOSE ISOMERASE INHIBITOR, D-THREONOHYDROXAMIC ACID, AND HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHIC STRUCTURE OF THE ENZYME-INHIBITOR COMPLEX | | Descriptor: | 2,3,4,N-TETRAHYDROXY-BUTYRIMIDIC ACID, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-09-01 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, and Characterization of a Potent Xylose Isomerase Inhibitor, D-Threonohydroxamic Acid, and High-Resolution X-Ray Crystallographic Structure of the Enzyme-Inhibitor Complex
Biochemistry, 34, 1995
|
|
2H7Q
 
 | | Cytochrome P450cam complexed with imidazole | | Descriptor: | Cytochrome P450-cam, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Verras, A, Alian, A, Montellano, P.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome P450 active site plasticity: attenuation of imidazole binding in cytochrome P450cam by an L244A mutation.
Protein Eng.Des.Sel., 19, 2006
|
|
1THL
 
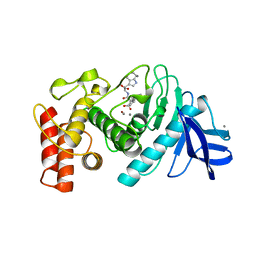 | | Thermolysin complexed with a novel glutaramide derivative, n-(1-(2(r,s)-carboxy-4-phenylbutyl) cyclopentylcarbonyl)-(s)-tryptophan | | Descriptor: | CALCIUM ION, N-({1-[(2S)-2-carboxy-4-phenylbutyl]cyclopentyl}carbonyl)-L-tryptophan, THERMOLYSIN, ... | | Authors: | Holland, D.R, Matthews, B.W. | | Deposit date: | 1993-11-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Thermolysin and Neutral Endopeptidase 24.11 By a Novel Glutaramide Derivative; X-Ray Structure Determination of the Thermolysin-Inhibitor Complex
Biochemistry, 33, 1994
|
|
7X1O
 
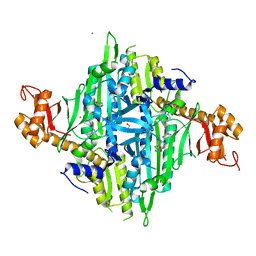 | |
4GPR
 
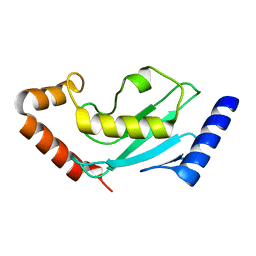 | |
4G1E
 
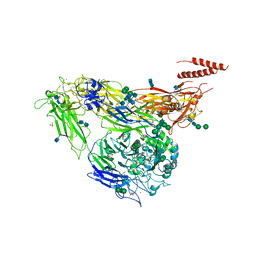 | | Crystal structure of integrin alpha V beta 3 with coil-coiled tag. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Mi, L, Zhu, J, Wang, W, Luo, B, Springer, T.A. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | AlphaV Beta3 Integrin Crystal Structures and their Functional Implications
Biochemistry, 51, 2012
|
|
4PW5
 
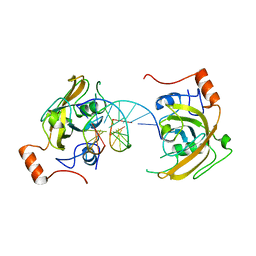 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex I | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
2YYK
 
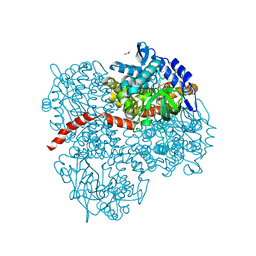 | | Crystal structure of the mutant of HpaB (T198I, A276G, and R466H) | | Descriptor: | 4-hydroxyphenylacetate-3-hydroxylase, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, S.-H, Hisano, T, Takeda, K, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-04-30 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Oxygenase Component (HpaB) of the 4-Hydroxyphenylacetate 3-Monooxygenase from Thermus thermophilus HB8
J.Biol.Chem., 282, 2007
|
|
1EY8
 
 | |
1EY7
 
 | |
3R1A
 
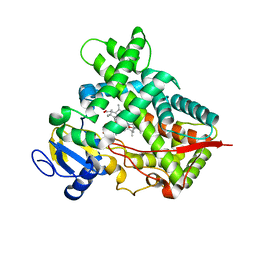 | | Closed crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
4TLM
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 2 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
5CNR
 
 | |
3GUZ
 
 | | Structural and substrate-binding studies of pantothenate synthenate (PS)provide insights into homotropic inhibition by pantoate in PS's | | Descriptor: | PANTOATE, Pantothenate synthetase | | Authors: | Chakrabarti, K.S, Thakur, K.G, Gopal, B, Sarma, S.P. | | Deposit date: | 2009-03-30 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray crystallographic and NMR studies of pantothenate synthetase provide insights into the mechanism of homotropic inhibition by pantoate
Febs J., 277, 2010
|
|
4TYO
 
 | | PPIase in complex with a non-phosphate small molecule inhibitor. | | Descriptor: | 3-(6-fluoro-1H-benzimidazol-2-yl)-N-(naphthalen-2-ylcarbonyl)-D-alanine, GLYCEROL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (III): Optimizing affinity beyond the phosphate recognition pocket.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5ZG2
 
 | | Crystal structure of the GluA2o LBD in complex with ZK200775 and Compound-2 | | Descriptor: | 9-(4-~{tert}-butylphenyl)-3,4-dihydropyrido[2,1-c][1,2,4]thiadiazine 2,2-dioxide, ACETATE ION, GLYCEROL, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
3GS4
 
 | | Human transthyretin (TTR) complexed with 3-(9H-fluoren-9-ylideneaminooxy)propanoic acid (inhibitor 15) | | Descriptor: | 3-[(9H-fluoren-9-ylideneamino)oxy]propanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis.
Plos One, 4, 2009
|
|
3CDE
 
 | | Crystal structure of HCV NS5B polymerase with a novel Pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-02-26 | | Release date: | 2009-03-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda(6)-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 3: Further optimization of the 2-, 6-, and 7'-substituents and initial pharmacokinetic assessments.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1BM0
 
 | | CRYSTAL STRUCTURE OF HUMAN SERUM ALBUMIN | | Descriptor: | SERUM ALBUMIN | | Authors: | Sugio, S, Kashima, A, Mochizuki, S, Noda, M, Kobayashi, K. | | Deposit date: | 1998-07-28 | | Release date: | 1999-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human serum albumin at 2.5 A resolution.
Protein Eng., 12, 1999
|
|
3ZS5
 
 | | Structural basis for kinase selectivity of three clinical p38alpha inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MITOGEN-ACTIVATED PROTEIN KINASE 14, ... | | Authors: | Azevedo, R, van Zeeland, M, Raaijmakers, H.C.A, Kazemier, B, Oubrie, A. | | Deposit date: | 2011-06-23 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure of p38 alpha bound to TAK-715: comparison with three classic inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
1RI4
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYLMETHIONINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RQY
 
 | | 9-amino-[N-(2-dimethylamino)proply]-acridine-4-carboxamide bound to d(CGTACG)2 | | Descriptor: | 5'-D(CP*GP*TP*AP*CP*G)-3', 9-AMINO-N-[3-(DIMETHYLAMINO)PROPYL]ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, ... | | Authors: | Adams, A, Guss, J.M, Denny, W.A, Wakelin, L.P. | | Deposit date: | 2003-12-07 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of 9-amino-[N-(2-dimethylamino)propyl]acridine-4-carboxamide bound to d(CGTACG)(2): a comparison of structures of d(CGTACG)(2) complexed with intercalatorsin the presence of cobalt.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1G0Y
 
 | | IL-1 RECEPTOR TYPE 1 COMPLEXED WITH ANTAGONIST PEPTIDE AF10847 | | Descriptor: | ANTAGONIST PEPTIDE AF10847, INTERLEUKIN-1 RECEPTOR, TYPE I | | Authors: | Vigers, G.P.A, Dripps, D.J, Edwards, C.K, Brandhuber, B.J. | | Deposit date: | 2000-10-09 | | Release date: | 2000-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of a small antagonist peptide bound to interleukin-1 receptor type 1.
J.Biol.Chem., 275, 2000
|
|
