5ZSY
 
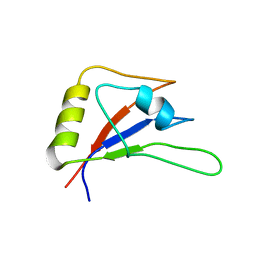 | | RBM10-RRM2 domain and its lung cancer related mutant | | Descriptor: | RNA-binding protein 10 | | Authors: | Qin, H. | | Deposit date: | 2018-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural, dynamics, and RNA binding comparison between RBM10-RRM2 domain and its lung cancer related mutation.
To be published
|
|
3EX6
 
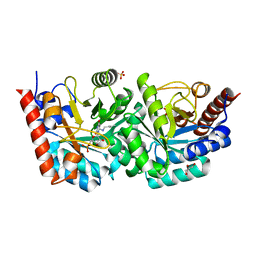 | | D312N mutant of human orotidyl-5'-monophosphate decarboxylase in complex with 6-azido-UMP, covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, Orotidine-5'-phosphate decarboxylase, ... | | Authors: | Heinrich, D, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
2DEF
 
 | |
6A63
 
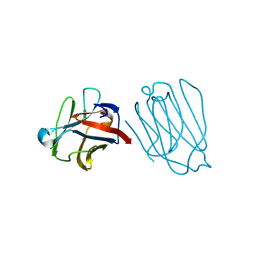 | | Placental protein 13/galectin-13 variant R53HH57R with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A66
 
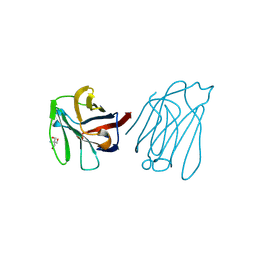 | | Placental protein 13/galectin-13 variant R53H with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J.Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
1FAP
 
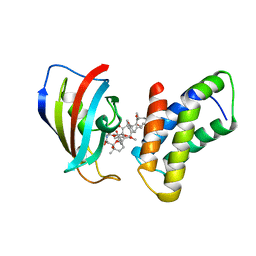 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | FK506-BINDING PROTEIN, FRAP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Choi, J, Chen, J, Schreiber, S.L, Clardy, J. | | Deposit date: | 1996-03-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP.
Science, 273, 1996
|
|
8AF8
 
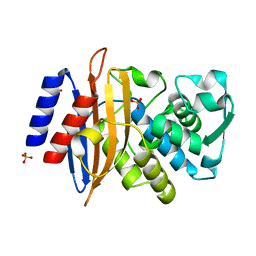 | | Room temperature SSX crystal structure of CTX-M-14 (5K dataset) | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Oberthuer, D, Perbandt, M, Prester, A, Rohde, H, Betzel, C, Yefanov, O. | | Deposit date: | 2022-07-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
5IEP
 
 | | Structure of CDL2.3b, a computationally designed Vitamin-D3 binder | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.3b | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5OOM
 
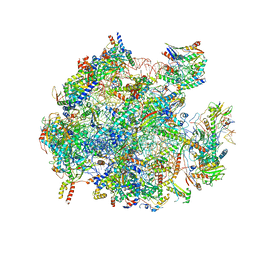 | | Structure of a native assembly intermediate of the human mitochondrial ribosome with unfolded interfacial rRNA | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Brown, A, Rathore, S, Kimanius, D, Aibara, S, Bai, X.C, Rorbach, J, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-13 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of the human mitochondrial ribosome in native states of assembly.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2NP8
 
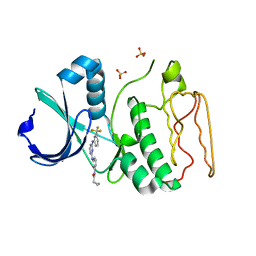 | | Structural Basis for the Inhibition of Aurora A Kinase by a Novel Class of High Affinity Disubstituted Pyrimidine Inhibitors | | Descriptor: | N-{3-[(4-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}PYRIMIDIN-2-YL)AMINO]PHENYL}CYCLOPROPANECARBOXAMIDE, SULFATE ION, Serine/threonine-protein kinase 6 | | Authors: | Tari, L.W, Hoffman, I.D, Bensen, D.C, Hunter, M.J, Nix, J, Nelson, K.J, McRee, D.E, Swanson, R.V. | | Deposit date: | 2006-10-26 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the inhibition of Aurora A kinase by a novel class of high affinity disubstituted pyrimidine inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2N4X
 
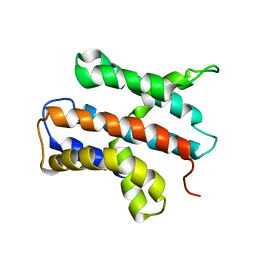 | |
5IIR
 
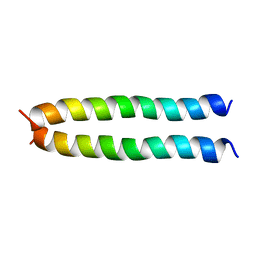 | |
6OQB
 
 | | Co-crystal structure of Mcl1 with inhibitor 10 | | Descriptor: | (4S,7aR,9aR,10S,11E,15R)-6'-chloro-15-ethyl-10-hydroxy-3',4',7a,8,9,9a,10,13,14,15-decahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)-16lambda~6~-cyclobuta[i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-4,1'-naphthalene]-16,16,18(7H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6S2O
 
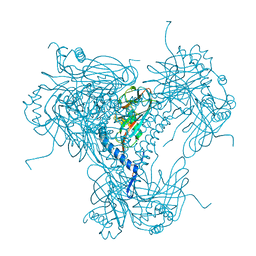 | |
5I6K
 
 | |
5I7O
 
 | |
3FFA
 
 | | Crystal Structure of a fast activating G protein mutant | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Chauhan, R, Kapoor, N. | | Deposit date: | 2008-12-02 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence for a sequential release mechanism for activation of heterotrimeric g proteins.
J.Mol.Biol., 393, 2009
|
|
5I9Z
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with danusertib (PHA739358) | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5IOV
 
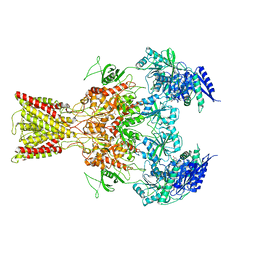 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine/Ro25-6981-bound conformation | | Descriptor: | 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IPV
 
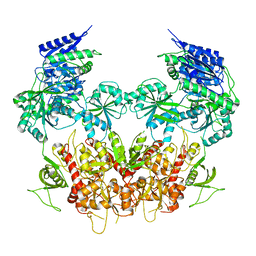 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 1 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, McHaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.25 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IAK
 
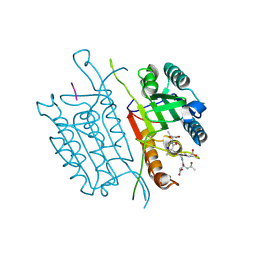 | | Caspase 3 V266S | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, SODIUM ION | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IQU
 
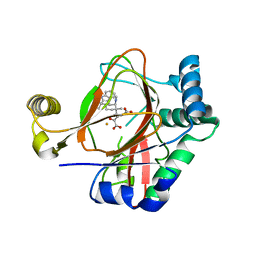 | | WelO5 G166D variant bound to Fe(II), 2-oxoglutarate, and 12-epifischerindole U | | Descriptor: | (6aS,9R,10R,10aS)-9-ethyl-10-isocyano-6,6,9-trimethyl-5,6,6a,7,8,9,10,10a-octahydroindeno[2,1-b]indole, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Mitchell, A.J, Maggiolo, A.O, Boal, A.K. | | Deposit date: | 2016-03-11 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5.
Nat.Chem.Biol., 12, 2016
|
|
6O6G
 
 | | Co-crystal structure of Mcl1 with inhibitor | | Descriptor: | (3S)-5-(cyclobutylmethyl)-3-(2,4-dichlorophenyl)-2,3,4,5-tetrahydro-1,5-benzoxazepine-7-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-03-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
5IA4
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with foretinib (XL880) | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5IAN
 
 | | Caspase 3 V266N | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, LEU-SER-SER, ... | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
