1YSM
 
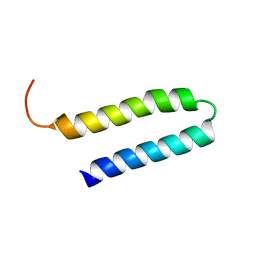 | | NMR Structure of N-terminal domain (Residues 1-77) of Siah-Interacting Protein. | | Descriptor: | Calcyclin-binding protein | | Authors: | Bhattacharya, S, Lee, Y.T, Michowski, W, Jastrzebska, B, Filipek, A, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Modular Structure of SIP Facilitates Its Role in Stabilizing Multiprotein Assemblies.
Biochemistry, 44, 2005
|
|
5I7R
 
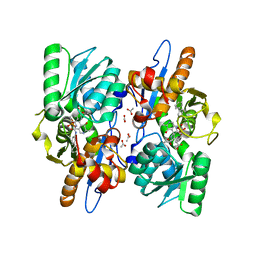 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 2 [3-(3-([1,1'-biphenyl]-3-yl)ureido)benzoic acid] | | Descriptor: | 3-{[([1,1'-biphenyl]-3-yl)carbamoyl]amino}benzoic acid, ACETATE ION, O-phosphoserine sulfhydrylase, ... | | Authors: | Schnell, R, Maric, S, Schneider, G. | | Deposit date: | 2016-02-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
5I0S
 
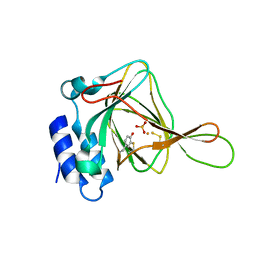 | | Thiosulfate bound Cysteine Dioxygenase at pH 6.2 | | Descriptor: | Cysteine dioxygenase type 1, FE (III) ION, THIOSULFATE | | Authors: | Kean, K.M, Driggers, C.M, Karplus, P.A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Insights into the Role of the Cys-Tyr Crosslink and Inhibitor Recognition by Mammalian Cysteine Dioxygenase.
J. Mol. Biol., 428, 2016
|
|
6A65
 
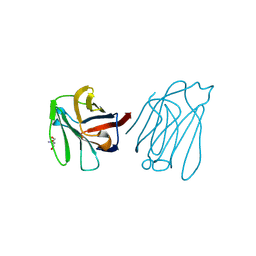 | | Placental protein 13/galectin-13 variant R53HR55N with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
5I6O
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 20.70 MGy | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
5I9S
 
 | | MicroED structure of proteinase K at 1.75 A resolution | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Hattne, J, Shi, D, de la Cruz, M.J, Reyes, F.E, Gonen, T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.75 Å) | | Cite: | Modeling truncated pixel values of faint reflections in MicroED images.
J.Appl.Crystallogr., 49, 2016
|
|
5I7H
 
 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 6 [3-(3-(4-Bromophenyl)ureido)benzoic acid] | | Descriptor: | 3-{[(4-bromophenyl)carbamoyl]amino}benzoic acid, CHLORIDE ION, O-phosphoserine sulfhydrylase, ... | | Authors: | Schnell, R, Maric, S, Schneider, G. | | Deposit date: | 2016-02-17 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
5IA1
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with MLN8054 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[9-CHLORO-7-(2,6-DIFLUOROPHENYL)-5H-PYRIMIDO[5,4-D][2]BENZAZEPIN-2-YL]AMINO}BENZOIC ACID, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5ZUZ
 
 | |
5IAB
 
 | | Caspase 3 V266D | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASK, Caspase-3 | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IBU
 
 | | 6652 Fab (unbound) | | Descriptor: | 6652 Heavy Chain, 6652 Light Chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain.
Nat. Med., 22, 2016
|
|
5I9Y
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with dasatinib | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.228 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5IEN
 
 | | Structure of CDL2.2, a computationally designed Vitamin-D3 binder | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.2, GLYCEROL | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5IF9
 
 | |
3EF7
 
 | |
1Z5B
 
 | |
5I8A
 
 | | TrkA with (6~{R})-3-methylsulfanyl-6-phenyl-1-(1~{H}-pyrazol-3-yl)-6,7-dihydro-5~{H}-thieno[3,4-c]pyridin-4-one | | Descriptor: | (6R)-3-(methylsulfanyl)-6-phenyl-1-(1H-pyrazol-3-yl)-6,7-dihydrothieno[3,4-c]pyridin-4(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Su, H.P. | | Deposit date: | 2016-02-18 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Potent, selective and orally bioavailable leucine-rich repeat kinase 2 (LRRK2) inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
2N40
 
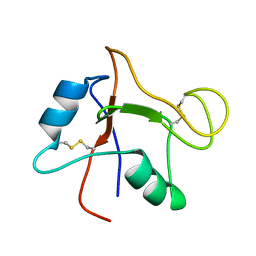 | |
5NRL
 
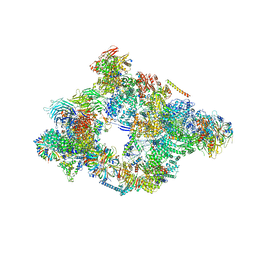 | | Structure of a pre-catalytic spliceosome. | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | Authors: | Plaschka, C, Lin, P.-C, Nagai, K. | | Deposit date: | 2017-04-24 | | Release date: | 2017-05-31 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure of a pre-catalytic spliceosome.
Nature, 546, 2017
|
|
5I4Q
 
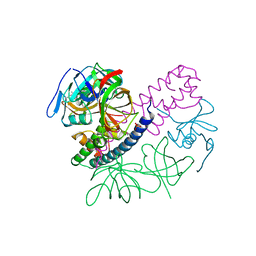 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (domains 2 and 3) | | Descriptor: | CHLORIDE ION, Contact-dependent inhibitor A, Contact-dependent inhibitor I, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
3EHS
 
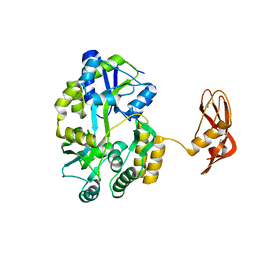 | |
5I6P
 
 | |
5I9B
 
 | | Caspase 3 V266A | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, SODIUM ION | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6A18
 
 | | Crystal structure of CYP90B1 in complex with 1,6-hexandiol | | Descriptor: | CHLORIDE ION, Cytochrome P450 90B1, GLYCEROL, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
5I9X
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with bosutinib (SKI-606) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.427 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
