2XQN
 
 | | Complex of the 2nd and 3rd LIM domains of TES with the EVH1 DOMAIN of MENA and the N-Terminal domain of actin-like protein Arp7A | | Descriptor: | ACTIN-LIKE PROTEIN 7A, ENABLED HOMOLOG, TESTIN, ... | | Authors: | Knowles, P.P, Briggs, D.C, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2010-09-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Molecular recognition of the Tes LIM2-3 domains by the actin-related protein Arp7A.
J. Biol. Chem., 286, 2011
|
|
2IJH
 
 | | Crystal structure analysis of ColE1 ROM mutant F14W | | Descriptor: | Regulatory protein rop | | Authors: | Ladner, J.E. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
5VC0
 
 | | Crystal structure of human CYP3A4 bound to ritonavir | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, RITONAVIR | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-03-30 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | High-Level Production and Properties of the Cysteine-Depleted Cytochrome P450 3A4.
Biochemistry, 56, 2017
|
|
5VCE
 
 | |
5KPK
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0209 | | Descriptor: | (4~{S})-3-cyclopropyl-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
9CRS
 
 | | Native human GABAA receptor of beta2-alpha1-beta2-alpha1-gamma2 assembly | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, J, Hibbs, R.E, Noviello, C.M. | | Deposit date: | 2024-07-22 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Resolving native GABA A receptor structures from the human brain.
Nature, 638, 2025
|
|
9CTP
 
 | | Native human GABAA receptor of beta2-alpha1-beta2-alpha3-gamma2 assembly | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, J, Hibbs, R.E, Noviello, C.M. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Resolving native GABA A receptor structures from the human brain.
Nature, 638, 2025
|
|
8BVL
 
 | |
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3OKO
 
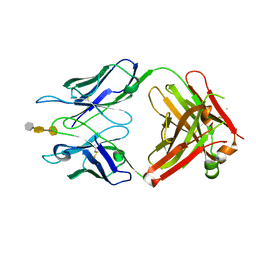 | | Crystal structure of S25-39 in complex with Kdo(2.8)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
6GD7
 
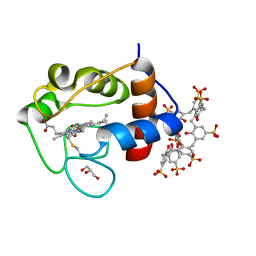 | | Cytochrome c in complex with Sulfonato-calix[8]arene, H3 form with PEG | | Descriptor: | Cytochrome c iso-1, GLYCEROL, HEME C, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1YSO
 
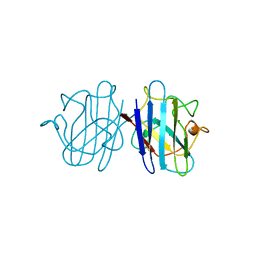 | | YEAST CU, ZN SUPEROXIDE DISMUTASE WITH THE REDUCED BRIDGE BROKEN | | Descriptor: | COPPER (I) ION, YEAST CU, ZN SUPEROXIDE DISMUTASE, ... | | Authors: | Parge, H.E, Crane, B.R, Tsang, J, Tainer, J.A. | | Deposit date: | 1995-12-21 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase.
Biochemistry, 35, 1996
|
|
1YPO
 
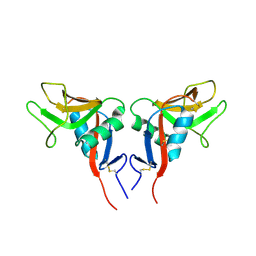 | |
6FS2
 
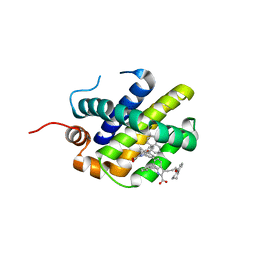 | | MCL1 in complex with indole acid ligand | | Descriptor: | 7-(2-methylphenyl)-3-[3-(5,6,7,8-tetrahydronaphthalen-1-yloxy)propyl]-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
1REK
 
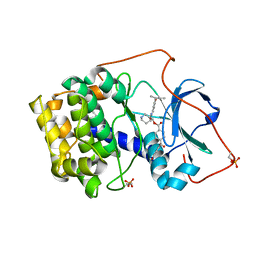 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 8 | | Descriptor: | 3-[(3-SEC-BUTYL-4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-(2-HYDROXY-5-METHOXYBENZOYL)BENZOATE, PENTANAL, cAMP-dependent protein kinase, ... | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
4MRQ
 
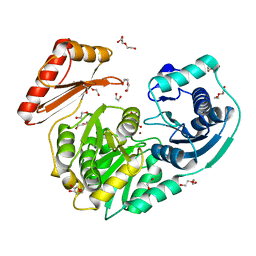 | | Crystal Structure of wild-type unphosphorylated PMM/PGM | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, ... | | Authors: | Lee, Y, Beamer, L. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Promotion of enzyme flexibility by dephosphorylation and coupling to the catalytic mechanism of a phosphohexomutase.
J.Biol.Chem., 289, 2014
|
|
3T5Z
 
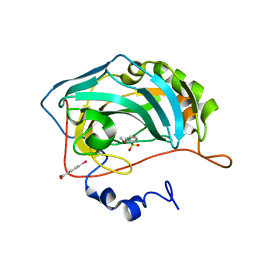 | | Crystal structure of the human carbonic anhydrase II in complex with N-methoxy-benzenesulfonamide | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, N-methoxybenzenesulfonamide, ... | | Authors: | Di Fiore, A, Maresca, A, Alterio, V, Supuran, C.T, De Simone, G. | | Deposit date: | 2011-07-28 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray crystallographic studies for the binding of N-substituted benzenesulfonamides to human isoform II.
Chem.Commun.(Camb.), 47, 2011
|
|
2JKW
 
 | | Pseudoazurin M16F | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Yanagisawa, S, Crowley, P.B, Firbank, S.J, Lawler, A.T, Hunter, D.M, McFarlane, W, Li, C, Kohzuma, T, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pi-Interaction Tuning of the Active Site Properties of Metalloproteins.
J.Am.Chem.Soc., 130, 2008
|
|
5VNZ
 
 | | Structure of a TRAF6-Ubc13~Ub complex | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Middleton, A.J, Day, C.L. | | Deposit date: | 2017-05-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1.
Nat Commun, 8, 2017
|
|
3AL0
 
 | | Crystal structure of the glutamine transamidosome from Thermotoga maritima in the glutamylation state. | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Glutamyl-tRNA(Gln) amidotransferase subunit A, Glutamyl-tRNA(Gln) amidotransferase subunit C,Linker,Glutamate--tRNA ligase 2, ... | | Authors: | Ito, T, Yokoyama, S. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.368 Å) | | Cite: | Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions.
Nature, 467, 2010
|
|
3SDP
 
 | |
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
5M96
 
 | |
2EUD
 
 | | Structures of Yeast Ribonucleotide Reductase I complexed with Ligands and Subunit Peptides | | Descriptor: | GEMCITABINE DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Dealwis, C, Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I define gemcitabine diphosphate binding and subunit assembly.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
