2QJW
 
 | |
2NW9
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferrous heme and 6-fluoro-tryptophan. Northeast Structural Genomics Target XcR13 | | Descriptor: | 6-FLUORO-L-TRYPTOPHAN, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Bruckmann, C, Thackray, S.J, Seetharaman, J, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
6BRD
 
 | | Crystal structure of rifampin monooxygenase from Streptomyces venezuelae, complexed with rifampin and FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Cox, G, Kelso, J, Stogios, P.J, Savchenko, A, Anderson, W.F, Wright, G.D, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-30 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action.
Cell Chem Biol, 25, 2018
|
|
3FWJ
 
 | | Ferric camphor bound Cytochrome P450cam containing a selenocysteine as the 5th heme ligand, orthorombic crystal form | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Schlichting, I, von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FWI
 
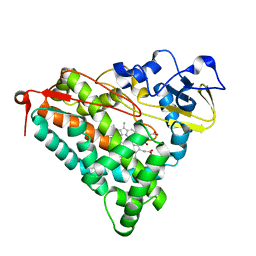 | | Ferric camphor bound Cytochrome P450cam containing a selenocysteine as the 5th heme ligand, tetragonal crystal form | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Schlichting, I, von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8VG9
 
 | |
4AKD
 
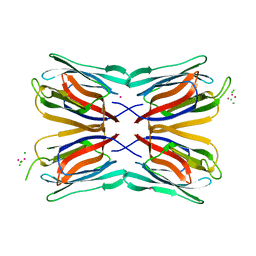 | | High resolution structure of Mannose Binding lectin from Champedak (CMB) | | Descriptor: | CADMIUM ION, CHLORIDE ION, MANNOSE-SPECIFIC LECTIN KM+ | | Authors: | Gabrielsen, M, Abdul-Rahman, P.S, Othman, S, Hashim, O.H, Cogdell, R.J. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and Binding Specificity of Galactose- and Mannose-Binding Lectins from Champedak: Differences from Jackfruit Lectins
Acta Crystallogr.,Sect.F, 70, 2014
|
|
9DTS
 
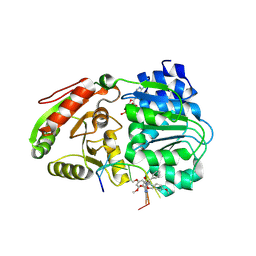 | | Crystal structure of the human eIF4A1/AMPPNP/amidino-rocaglate/polypurine RNA complex | | Descriptor: | (3aR,4R,5S,5aR,10bR)-3a-hydroxy-N,8,10-trimethoxy-5a-(4-methoxyphenyl)-N,2-dimethyl-5-phenyl-3a,4,5,5a-tetrahydro-1H-[1]benzofuro[3',2':1,5]cyclopenta[1,2-d]imidazole-4-carboxamide, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Conley, J.F, Allen, K.N. | | Deposit date: | 2024-10-01 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Basis for the Improved RNA Clamping of Amidino-Rocaglates to eIF4A1.
Acs Omega, 10, 2025
|
|
4GXS
 
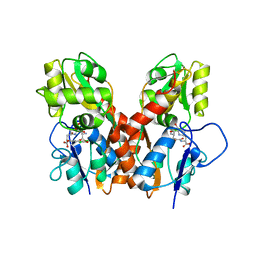 | | Ligand binding domain of GluA2 (AMPA/glutamate receptor) bound to (-)-kaitocephalin | | Descriptor: | (5R)-2-[(1S,2R)-2-amino-2-carboxy-1-hydroxyethyl]-5-{(2S)-2-carboxy-2-[(3,5-dichloro-4-hydroxybenzoyl)amino]ethyl}-L-proline, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9634 Å) | | Cite: | The structure of (-)-kaitocephalin bound to the ligand binding domain of the (S)-alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)/glutamate receptor, GluA2.
J.Biol.Chem., 287, 2012
|
|
2NW8
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferrous heme and tryptophan. Northeast Structural Genomics Target XcR13. | | Descriptor: | MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, ... | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Bruckmann, C, Thackray, S.J, Seetharaman, J, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4K3Q
 
 | | E. coli sliding clamp in complex with AcQLDAF | | Descriptor: | (ACE)QLDAF, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
3I0Y
 
 | |
4K3M
 
 | | E.coli sliding clamp in complex with AcALDLF peptide | | Descriptor: | ALDLF, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
6S4G
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruggieri, F, Campillo Brocal, J.C, Humble, M.S, Walse, B, Logan, D.T, Berglund, P. | | Deposit date: | 2019-06-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Insight into the dimer dissociation process of the Chromobacterium violaceum (S)-selective amine transaminase.
Sci Rep, 9, 2019
|
|
4K3O
 
 | | E. coli sliding clamp in complex with AcQADLF | | Descriptor: | (ACE)QADLF, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4K3K
 
 | |
4K3P
 
 | | E. coli sliding clamp in complex with AcQLALF | | Descriptor: | (ACE)QLALF, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4K3R
 
 | | E. coli sliding clamp in complex with AcQLDLA | | Descriptor: | (ACE)QLDLA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4K3L
 
 | | E. coli sliding clamp in complex with AcLF dipeptide | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
3H06
 
 | |
4K3S
 
 | | E. coli sliding clamp in P1 crystal space group | | Descriptor: | CALCIUM ION, DNA polymerase III subunit beta, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
1MMA
 
 | | X-RAY STRUCTURES OF THE MGADP, MGATPGAMMAS, AND MGAMPPNP COMPLEXES OF THE DICTYOSTELIUM DISCOIDEUM MYOSIN MOTOR DOMAIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Rayment, I. | | Deposit date: | 1997-07-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain.
Biochemistry, 36, 1997
|
|
4JUU
 
 | | Crystal structure of a putative hydroxyproline epimerase from xanthomonas campestris (TARGET EFI-506516) with bound phosphate and unknown ligand | | Descriptor: | CHLORIDE ION, Epimerase, PHOSPHATE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-25 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative hydroxyproline epimerase from xanthomonas campestris (TARGET EFI-506516) with bound phosphate and unknown ligand
To be Published
|
|
1MMG
 
 | | X-RAY STRUCTURES OF THE MGADP, MGATPGAMMAS, AND MGAMPPNP COMPLEXES OF THE DICTYOSTELIUM DISCOIDEUM MYOSIN MOTOR DOMAIN | | Descriptor: | MAGNESIUM ION, MYOSIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Rayment, I. | | Deposit date: | 1997-07-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain.
Biochemistry, 36, 1997
|
|
