6HJS
 
 | | Crystal structure of glutathione transferase Omega 1C from Trametes versicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE, Uncharacterized protein | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-09-04 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 2021
|
|
7AYC
 
 | | Crystal Structure of human mitochondrial 2-Enoyl Thioester Reductase (MECR) with single mutation G165Q | | Descriptor: | CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, mitochondrial | | Authors: | Rahman, M.T, Koski, M.K, Autio, K.J, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2020-11-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An engineered variant of MECR reductase reveals indispensability of long-chain acyl-ACPs for mitochondrial respiration.
Nat Commun, 14, 2023
|
|
6DFI
 
 | |
7AYZ
 
 | |
7PGO
 
 | | Crystal Structure of a Class D Carbapenemase_R250A | | Descriptor: | 1-BUTANOL, BROMIDE ION, Beta-lactamase | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
6HKN
 
 | | Crystal structure of Compound 35 with ERK5 | | Descriptor: | Mitogen-activated protein kinase 7, [2-azanyl-4-(trifluoromethyloxy)phenyl]-[4-(7-methoxyquinazolin-4-yl)piperidin-1-yl]methanone | | Authors: | Nguyen, D, Lemos, C, Wortmann, L, Eis, K, Holton, S.J, Boemer, U, Lechner, C, Prechtl, S, Suelze, D, Siegel, F, Prinz, F, Lesche, R, Nicke, B, Mumberg, D, Bauser, M, Haegebarth, A. | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective (Piperidin-4-yl)pyrido[3,2- d]pyrimidine Based in Vitro Probe BAY-885 for the Kinase ERK5.
J. Med. Chem., 62, 2019
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | Descriptor: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
6HKS
 
 | |
7PIZ
 
 | |
7AYY
 
 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
8TJK
 
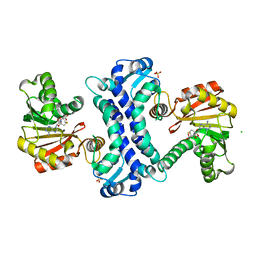 | | SAM-dependent methyltransferase RedM bound to SAH | | Descriptor: | CHLORIDE ION, RedM, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-07-22 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
7AZ0
 
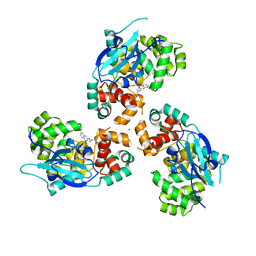 | |
8TUC
 
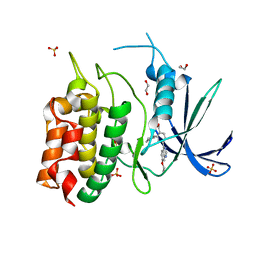 | | Unphosphorylated CaMKK2 in complex with CC-8977 | | Descriptor: | (4M)-2-cyclopentyl-4-(7-ethoxyquinazolin-4-yl)benzoic acid, 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Bernard, S.M, Shanmugasundaram, V, D'Agostino, L. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Small Molecule Inhibitors and Ligand Directed Degraders of Calcium/Calmodulin Dependent Protein Kinase Kinase 1 and 2 (CaMKK1/2).
J.Med.Chem., 66, 2023
|
|
8U1W
 
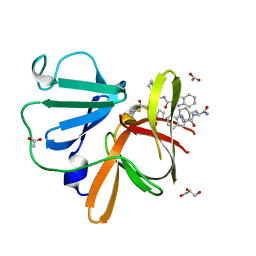 | | Structure of Norovirus (Hu/GII.4/Sydney/NSW0514/2012/AU) protease bound to inhibitor NV-004 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidase C37, ... | | Authors: | Eruera, A.R, Campbell, A.C, Krause, K.L. | | Deposit date: | 2023-09-03 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Inhibitor-Bound GII.4 Sydney 2012 Norovirus 3C-Like Protease.
Viruses, 15, 2023
|
|
7AVN
 
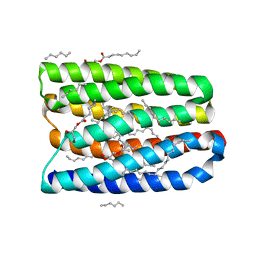 | | Structure of marine actinobacteria clade rhodopsin (MacR) in orange form in P1 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, ... | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Alternating Access Mechanism in a Minimalistic Light-Driven Proton Pump
To Be Published
|
|
6HM0
 
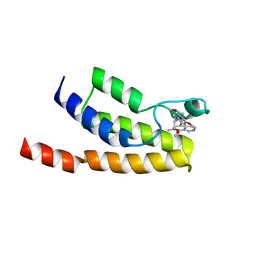 | | Crystal structure of human BRD9 bromodomain in complex with a PROTAC | | Descriptor: | (2~{S},4~{S})-1-[(2~{S})-2-[2-[2-[2-[4-[[2,6-dimethoxy-4-(2-methyl-1-oxidanylidene-2,7-naphthyridin-4-yl)phenyl]methyl]piperazin-1-yl]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 9 | | Authors: | Hughes, S.J, Zoppi, V, Ciulli, A. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Iterative Design and Optimization of Initially Inactive Proteolysis Targeting Chimeras (PROTACs) Identify VZ185 as a Potent, Fast, and Selective von Hippel-Lindau (VHL) Based Dual Degrader Probe of BRD9 and BRD7.
J.Med.Chem., 62, 2019
|
|
8TJI
 
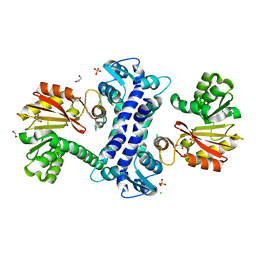 | | SAM-dependent methyltransferase RedM, apo | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, RedM, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-07-22 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
8AEI
 
 | |
6DPD
 
 | |
6HM4
 
 | | Crystal structure of Rad4 BRCT1,2 in complex with a Mdb1 phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA damage response protein Mdb1, ... | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.770186 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
8TGB
 
 | | Crystal structure of root lateral formation protein (RLF) b5-domain from Oryza sativa | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, root lateral formation protein (RLF) | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Benson, D.R. | | Deposit date: | 2023-07-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The N-terminal intrinsically disordered region of Ncb5or docks with the cytochrome b 5 core to form a helical motif that is of ancient origin.
Proteins, 92, 2024
|
|
7AYB
 
 | | Crystal Structure of wild type human mitochondrial 2-Enoyl Thioester Reductase (MECR) | | Descriptor: | ACETIC ACID, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, ... | | Authors: | Rahman, M.T, Koski, M.K, Autio, K.J, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2020-11-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An engineered variant of MECR reductase reveals indispensability of long-chain acyl-ACPs for mitochondrial respiration.
Nat Commun, 14, 2023
|
|
8TJJ
 
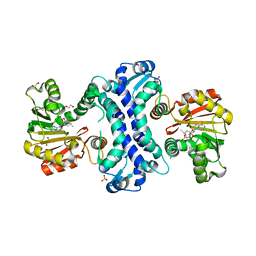 | | SAM-dependent methyltransferase RedM bound to SAM | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, RedM, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-07-22 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
7PFN
 
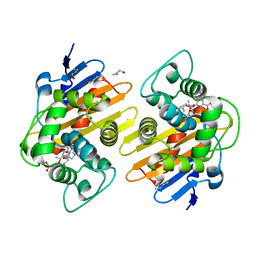 | |
6DQQ
 
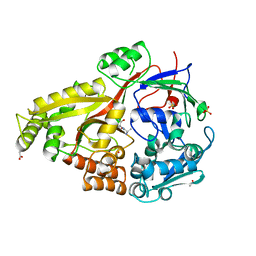 | |
