9ENB
 
 | |
9CVG
 
 | |
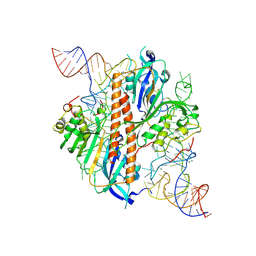
8ZR5
 
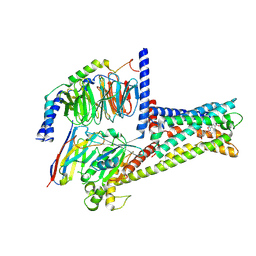 | | Cryo-EM Structure of GPR119-Gs-Firuglipel complex | | Descriptor: | 4-[5-[(1~{R})-1-(4-cyclopropylcarbonylphenoxy)propyl]-1,2,4-oxadiazol-3-yl]-2-fluoranyl-~{N}-[(2~{R})-1-oxidanylpropan-2-yl]benzamide, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Qiu, C, Du, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of GPR119-Gs complex
To Be Published
|
|
9B6E
 
 | | Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 | | Descriptor: | 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Park, C.-G, Zhang, F, Fedor, J, Feng, S, Suo, Y, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8.
Sci Adv, 10, 2024
|
|
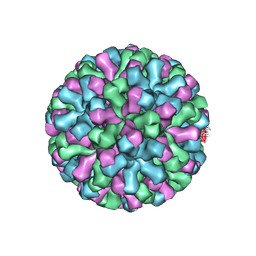
9EZ8
 
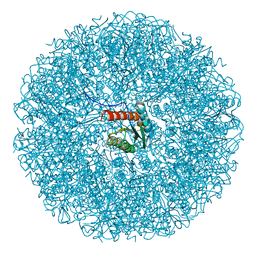 | | Cryo-EM structure of the icosahedral lumazine synthase from Vicia faba. | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Chee, M, Trapani, S, Hoh, F, Lai Kee Him, J, Yvon, M, Blanc, S, Bron, P. | | Deposit date: | 2024-04-11 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the icosahedral lumazine synthase from Vicia faba.
To Be Published
|
|
9B8P
 
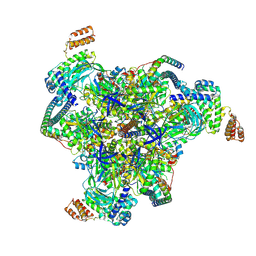 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, V1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H+-transporting V1 subunit D, H(+)-transporting two-sector ATPase, ... | | Authors: | Coupland, E.M, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
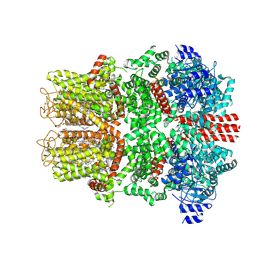
9CHR
 
 | |
8YTB
 
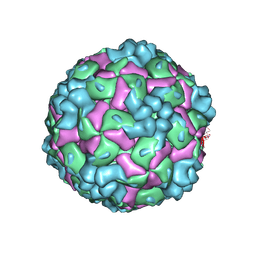 | | Cryo-EM structure of enterovirus A71 empty particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2024-03-25 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structure of enterovirus A71 empty particle
To Be Published
|
|
9C57
 
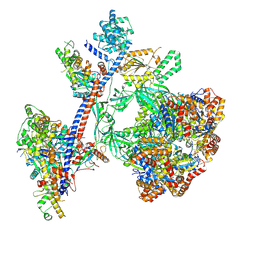 | | Reconstituted P400 Subcomplex of the human TIP60 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-05 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
8YXR
 
 | |
9AWK
 
 | | Bovine fetal muscle nAChR resting state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H, Hibbs, R.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Structural switch in acetylcholine receptors in developing muscle.
Nature, 632, 2024
|
|
8YTJ
 
 | | Cryo-EM structure of enterovirus A71 mature virion | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Huang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2024-03-26 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab h1A6.2
To Be Published
|
|
9EM8
 
 | | Oligomeric structure of SynDLP in presence of GDP | | Descriptor: | Slr0869 protein | | Authors: | Junglas, B, Gewehr, L, Schoennenbeck, P, Schneider, D, Sachse, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for GTPase activity and conformational changes of the bacterial dynamin-like protein SynDLP.
Cell Rep, 43, 2024
|
|
8ZFJ
 
 | | cryo-EM structure of GPR4-Gs complex at pH 8.5 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Ma, Y, Tang, M, Ru, H, Song, G. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | cryo-EM structure of GPR4-Gs complex at pH 8.5
to be published
|
|
8ZHE
 
 | | SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
9JA1
 
 | | The RNA polymerase II elongation complex from Saccharomyces cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*CP*TP*CP*CP*TP*TP*CP*TP*CP*CP*CP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*T)-3'), DNA (5'-D(P*TP*GP*GP*GP*AP*GP*AP*AP*GP*GP*AP*GP*C)-3'), ... | | Authors: | Yi, G, Ma, J, Zhang, P. | | Deposit date: | 2024-08-23 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The RNA polymerase II elongation complex from Saccharomyces cerevisiae
To Be Published
|
|
8ZHM
 
 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
8YW4
 
 | | Cryo-EM structure of the retatrutide-bound human GIPR-Gs complex | | Descriptor: | Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, W.Z, Zhou, Q.T, Cong, Z.T, Yuan, Q.N, Li, W.X, Zhao, F.H, Xu, H.E, Zhao, L.H, Yang, D.H, Wang, M.W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into the triple agonism at GLP-1R, GIPR and GCGR manifested by retatrutide.
Cell Discov, 10, 2024
|
|
9ENC
 
 | |
9CC3
 
 | | Human Mitochondrial LONP1 Stall State + casein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endogenous Co-purified substrate modeled as unknown residues, Lon protease homolog, ... | | Authors: | Mindrebo, J.T, Lander, G.C. | | Deposit date: | 2024-06-20 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural and mechanistic studies on human LONP1 redefine the hand-over-hand translocation mechanism.
Biorxiv, 2024
|
|
9CC0
 
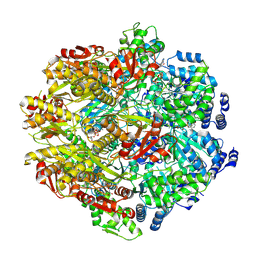 | |
9IJA
 
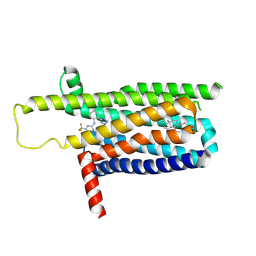 | | A local Cryo-EM structure of Bitter taste receptor TAS2R14 with Gi complex | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Taste receptor type 2 member 14 | | Authors: | Yuan, Q, Duan, J, Tao, L, Xu, E.H. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bitter taste receptor TAS2R14 activation and G protein assembly by an intracellular agonist.
Cell Res., 2024
|
|
8ZHH
 
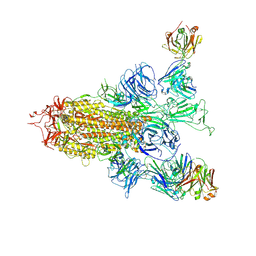 | | SARS-CoV-2 spike trimer (6P) in complex with two H18 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (5.55 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
8Z9X
 
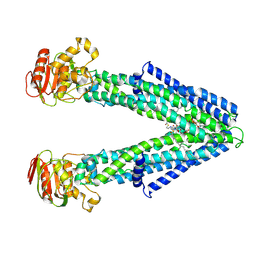 | |
9FJL
 
 | |
