8FFX
 
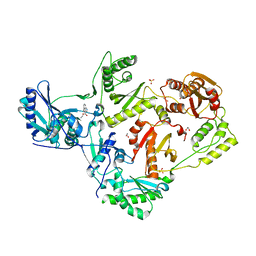 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 19980 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Reverse transcriptase/ribonuclease H, ... | | Authors: | Rumrill, S.R, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Targeting HIV-1 Reverse Transcriptase Using a Fragment-Based Approach.
Molecules, 28, 2023
|
|
8FE8
 
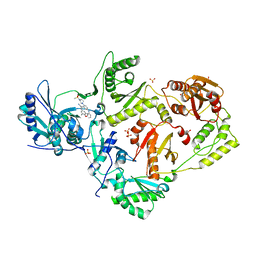 | | Crystal Structure of HIV-1 RT in Complex with the non-nucleoside inhibitor 18b1 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}pyrimidin-2-yl)amino]-2-[4-(methanesulfonyl)piperazin-1-yl]benzonitrile, Reverse transcriptase p51, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of diarylpyrimidine derivatives bearing piperazine sulfonyl as potent HIV-1 nonnucleoside reverse transcriptase inhibitors.
Commun Chem, 6, 2023
|
|
8GAP
 
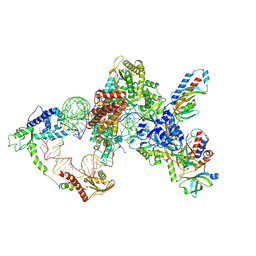 | | Structure of LARP7 protein p65-telomerase RNA complex in telomerase | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | Wang, Y, He, Y, Wang, Y, Yang, Y, Singh, M, Eichhorn, C.D, Zhou, Z.H, Feigon, J. | | Deposit date: | 2023-02-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of LARP7 Protein p65-telomerase RNA Complex in Telomerase Revealed by Cryo-EM and NMR.
J.Mol.Biol., 435, 2023
|
|
4NCG
 
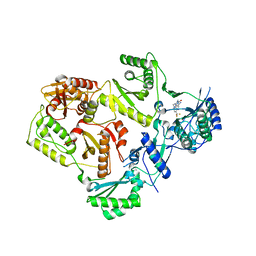 | | Discovery of Doravirine, an orally bioavailable non-nucleoside reverse transcriptase inhibitor potent against a wide range of resistant mutant HIV viruses | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of MK-1439, an orally bioavailable non-nucleoside reverse transcriptase inhibitor potent against a wide range of resistant mutant HIV viruses.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4O4G
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in complex with 4-((4-(mesitylamino)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ527), a non-nucleoside inhibitor | | Descriptor: | 4-({4-[(2,4,6-trimethylphenyl)amino]-1,3,5-triazin-2-yl}amino)benzonitrile, HIV-1 reverse transcriptase, p51 subunit, ... | | Authors: | Mislak, A.C, Frey, K.M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | A mechanistic and structural investigation of modified derivatives of the diaryltriazine class of NNRTIs targeting HIV-1 reverse transcriptase.
Biochim.Biophys.Acta, 1840, 2014
|
|
4O44
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in complex with 4-((4-(mesitylamino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ529), a non-nucleoside inhibitor | | Descriptor: | 4-({4-[3-(morpholin-4-yl)propoxy]-6-[(2,4,6-trimethylphenyl)amino]-1,3,5-triazin-2-yl}amino)benzonitrile, HIV-1 reverse transcriptase, p51 subunit, ... | | Authors: | Mislak, A.C, Frey, K.M, Anderson, K.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | A mechanistic and structural investigation of modified derivatives of the diaryltriazine class of NNRTIs targeting HIV-1 reverse transcriptase.
Biochim.Biophys.Acta, 1840, 2014
|
|
4PQU
 
 | | Crystal structure of HIV-1 Reverse Transcriptase in complex with RNA/DNA and dATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', ... | | Authors: | Das, K, Bandwar, R.P, Arnold, E. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
7SEP
 
 | |
7SJX
 
 | | Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein | | Descriptor: | Gag-Pol polyprotein | | Authors: | Lyumkis, D, Passos, D, Arnold, E, Harrison, J.J.E.K, Ruiz, F.X. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation.
Sci Adv, 8, 2022
|
|
4PUO
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*UP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-03-13 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
4Q0B
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with gap-RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-04-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
6X49
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ649), a Non-nucleoside Inhibitor | | Descriptor: | 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6X4F
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with methyl 2-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acetate (JLJ681), a Non-nucleoside Inhibitor | | Descriptor: | Reverse transcriptase/ribonuclease H, SULFATE ION, methyl (6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)acetate, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6WPJ
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and d4T | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA Primer 21-mer, DNA template 27-mer, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Post-Catalytic Complexes with Emtricitabine or Stavudine and HIV-1 Reverse Transcriptase Reveal New Mechanistic Insights for Nucleotide Incorporation and Drug Resistance.
Molecules, 25, 2020
|
|
6WPH
 
 | |
3FFI
 
 | | HIV-1 RT with pyridone non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-({6-[2-(3,4-dihydroisoquinolin-2(1H)-yl)-2-oxoethyl]-3-(dimethylamino)-2-oxo-1,2-dihydropyridin-4-yl}oxy)benzonitrile, RT p51, Reverse transcriptase/ribonuclease H | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyridone Diaryl Ether Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase
To be Published
|
|
6X4B
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-5-fluoro-8-methyl-2-naphthonitrile (JLJ655), a Non-nucleoside Inhibitor | | Descriptor: | 7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-5-fluoro-8-methylnaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6KDO
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y/M184V/F160M:DNA:lamivudine 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDK
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
3HVT
 
 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | Deposit date: | 1994-07-25 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
2WOM
 
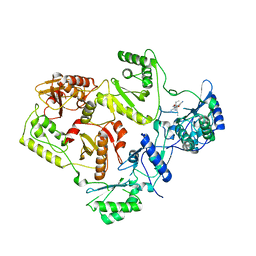 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (K103N). | | Descriptor: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | Deposit date: | 2009-07-27 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Lersivirine, a nonnucleoside reverse transcriptase inhibitor with activity against drug-resistant human immunodeficiency virus type 1.
Antimicrob. Agents Chemother., 54, 2010
|
|
6X4A
 
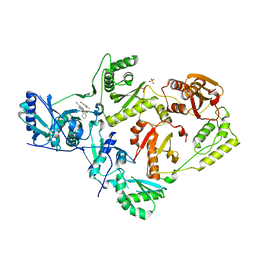 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ651), a Non-nucleoside Inhibitor | | Descriptor: | 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
3I0R
 
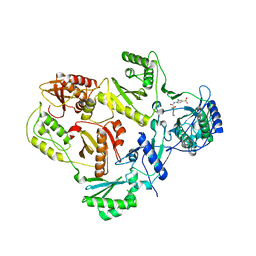 | | crystal structure of HIV reverse transcriptase in complex with inhibitor 3 | | Descriptor: | Reverse transcriptase/ribonuclease H, S-{2-[(2-chloro-4-sulfamoylphenyl)amino]-2-oxoethyl} 6-methyl-3,4-dihydroquinoline-1(2H)-carbothioate, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Substituted tetrahydroquinolines as potent allosteric inhibitors of reverse transcriptase and its key mutants.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6WPF
 
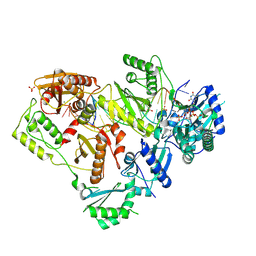 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and d4T | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA Primer 21-mer, DNA template 27-mer, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Post-Catalytic Complexes with Emtricitabine or Stavudine and HIV-1 Reverse Transcriptase Reveal New Mechanistic Insights for Nucleotide Incorporation and Drug Resistance.
Molecules, 25, 2020
|
|
5HBM
 
 | | Crystal Structure of a Dihydroxycoumarin RNase H Active-Site Inhibitor in Complex with HIV-1 Reverse Transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MANGANESE (II) ION, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.043 Å) | | Cite: | Crystal Structure of a Dihydroxycoumarin RNase H Active-Site Inhibitor in Complex with HIV-1 Reverse Transcriptase
To Be Published
|
|
