8C1Q
 
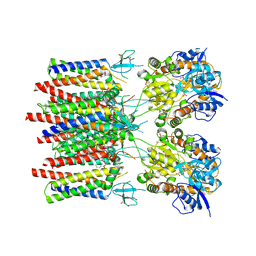 | | Resting state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
4NF5
 
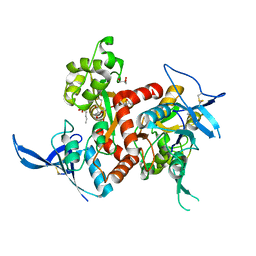 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and D-AP5 | | Descriptor: | 5-phosphono-D-norvaline, GLYCEROL, GLYCINE, ... | | Authors: | Jespersen, A, Tajima, N, Furukawa, H. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
4FAT
 
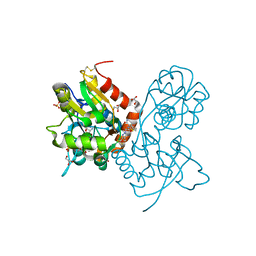 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-({[4-(hydroxymethyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]acetyl}amino)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Maclean, J.K.F, Kazemier, B. | | Deposit date: | 2012-05-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Functional analysis of a novel positive allosteric modulator of AMPA receptors derived from a structure-based drug design strategy.
Neuropharmacology, 64, 2013
|
|
8F0O
 
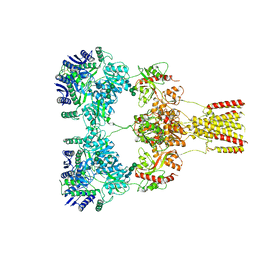 | |
4LZ5
 
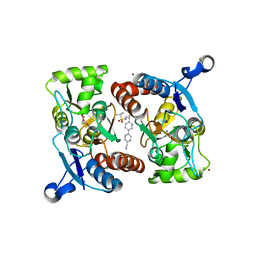 | |
8FWT
 
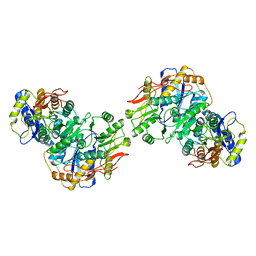 | | Structure of the amino terminal domain of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and competitive antagonist DNQX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Yen, L.Y, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWQ
 
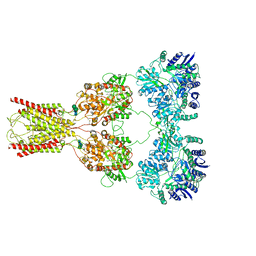 | | Structure of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWW
 
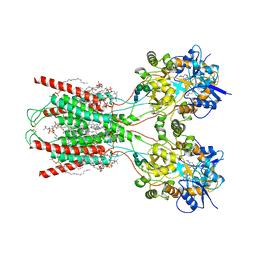 | | Structure of the ligand-binding and transmembrane domains of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and noncompetitive inhibitor perampanel | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
4E0W
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | Kainate induces various domain closures in AMPA and kainate receptors.
Neurochem Int, 61, 2012
|
|
8FWR
 
 | | Structure of the amino-terminal domain of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWV
 
 | | Structure of the amino-terminal domain of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and noncompetitive inhibitor perampanel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWU
 
 | | Structure of the ligand-binding and transmembrane domains of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and competitive antagonist DNQX | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yen, L.Y, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
4O3A
 
 | | Crystal structure of the glua2 ligand-binding domain in complex with L-aspartate at 1.80 a resolution | | Descriptor: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Krintel, C, Frydenvang, F, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4O3B
 
 | | Crystal structure of an open/closed glua2 ligand-binding domain dimer at 1.91 A resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krintel, C, de Rabassa, A.C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4O3C
 
 | | Crystal structure of the GLUA2 ligand-binding domain in complex with L-aspartate at 1.50 A resolution | | Descriptor: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Krintel, C, Frydenvang, K, Kaern, A.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4NF6
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and PPDA | | Descriptor: | (2S,3R)-1-(phenanthren-2-ylcarbonyl)piperazine-2,3-dicarboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Jespersen, A, Tajima, N, Furukawa, H. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
8FWS
 
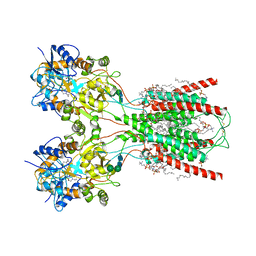 | | Structure of the ligand-binding and transmembrane domains of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
4NF8
 
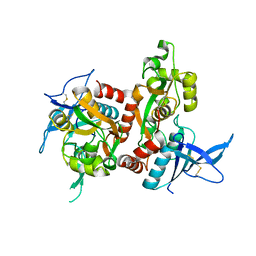 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and glutamate in PEG2000MME | | Descriptor: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Jespersen, A, Tajima, N, Furukawa, H. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.856 Å) | | Cite: | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
4N07
 
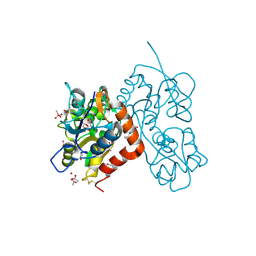 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-344 at 1.87 A resolution | | Descriptor: | 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | Authors: | Noerholm, A.B, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synthesis, pharmacological and structural characterization, and thermodynamic aspects of GluA2-positive allosteric modulators with a 3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide scaffold.
J.Med.Chem., 56, 2013
|
|
4F2O
 
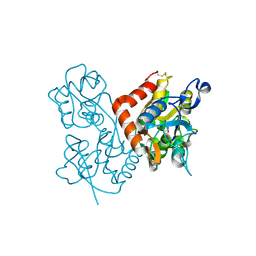 | | Quisqualate bound to the D655A mutant of the ligand binding domain of GluA3 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
4F3G
 
 | | Kainate bound to the ligand binding domain of GluA3i | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
4NWD
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | Descriptor: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
4NWC
 
 | | Crystal structure of the GluK3 ligand-binding domain (S1S2) in complex with the agonist (2S,4R)-4-(3-Methoxy-3-oxopropyl)glutamic acid at 2.01 A resolution. | | Descriptor: | (2S,4R)-4-(3-Methoxy-3-oxopropyl) glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
4G8N
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist G8M | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]cyclobutanecarboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Kastrup, J.S, Frydenvang, K, Gajhede, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors.
J.Struct.Biol., 180, 2012
|
|
4F3B
 
 | |
