4EMT
 
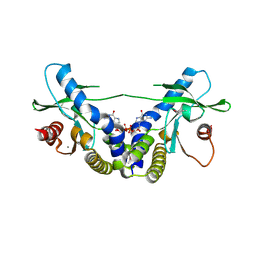 | | Crystal Structure of human STING bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Li, P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EMU
 
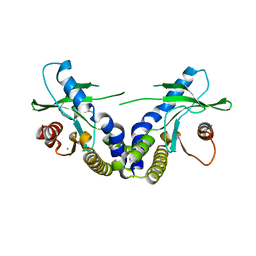 | | Crystal structure of ligand free human STING | | Descriptor: | CALCIUM ION, Transmembrane protein 173 | | Authors: | Li, P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1BL5
 
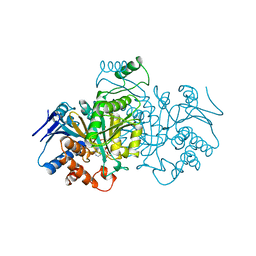 | | ISOCITRATE DEHYDROGENASE FROM E. COLI SINGLE TURNOVER LAUE STRUCTURE OF RATE-LIMITED PRODUCT COMPLEX, 10 MSEC TIME RESOLUTION | | Descriptor: | 2-OXOGLUTARIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION, ... | | Authors: | Stoddard, B.L, Cohen, B, Brubaker, M, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1998-07-23 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Millisecond Laue structures of an enzyme-product complex using photocaged substrate analogs.
Nat.Struct.Biol., 5, 1998
|
|
6YIG
 
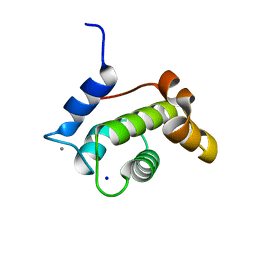 | | Crystal structure of the N-terminal EF-hand domain of Arabidopsis thaliana AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein, SODIUM ION | | Authors: | Yperman, K, Merceron, R, De Munck, S, Bloch, Y, Savvides, S.N, Pleskot, R, Van Damme, D. | | Deposit date: | 2020-04-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
6YEU
 
 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
6YET
 
 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
7LUF
 
 | | HSV1 polymerase ternary complex with dsDNA and PNU-183792 | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*CP*G)-3'), DNA polymerase, ... | | Authors: | Hayes, R.P, Klein, D. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural understanding of non-nucleoside inhibition in an elongating herpesvirus polymerase.
Nat Commun, 12, 2021
|
|
7BAJ
 
 | |
7BAK
 
 | |
7BAL
 
 | |
5V7V
 
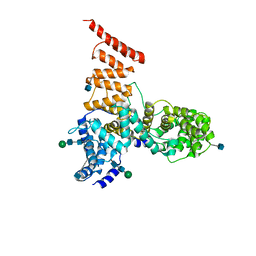 | | Cryo-EM structure of ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERAD-associated E3 ubiquitin-protein ligase component HRD3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mi, W, Schoebel, S, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-08-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
5V6P
 
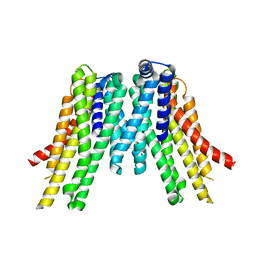 | | CryoEM structure of the ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Authors: | Schoebel, S, Mi, W, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
5I3U
 
 | |
4R5P
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor | | Descriptor: | 5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', 5'-D(*TP*GP*GP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3', HIV-1 reverse transcriptase, ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
