1IK6
 
 | |
1IK7
 
 | | Crystal Structure of the Uncomplexed Pelle Death Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PROBABLE SERINE/THREONINE-PROTEIN KINASE Pelle | | Authors: | Xiao, T, Gardner, K.H, Sprang, S.R. | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cosolvent-induced transformation
of a death domain tertiary structure
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IK8
 
 | | NMR structure of Alpha-Bungarotoxin | | Descriptor: | LONG NEUROTOXIN 1 | | Authors: | Niccolai, N, Ciutti, A, Spiga, O. | | Deposit date: | 2001-05-03 | | Release date: | 2001-05-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
1IK9
 
 | | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | | Descriptor: | DNA LIGASE IV, DNA REPAIR PROTEIN XRCC4 | | Authors: | Sibanda, B.L, Critchlow, S.E, Begun, J, Pei, X.Y, Jackson, S.P, Blundell, T.L, Pellegrini, L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an Xrcc4-DNA ligase IV complex.
Nat.Struct.Biol., 8, 2001
|
|
1IKA
 
 | |
1IKC
 
 | | NMR Structure of alpha-Bungarotoxin | | Descriptor: | long neurotoxin 1 | | Authors: | Niccolai, N, Spiga, O, Ciutti, A. | | Deposit date: | 2001-05-03 | | Release date: | 2001-05-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
1IKD
 
 | | ACCEPTOR STEM, NMR, 30 STRUCTURES | | Descriptor: | TRNA ALA ACCEPTOR STEM | | Authors: | Ramos, A, Varani, G. | | Deposit date: | 1996-11-15 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the acceptor stem of Escherichia coli tRNA Ala: role of the G3.U70 base pair in synthetase recognition.
Nucleic Acids Res., 25, 1997
|
|
1IKE
 
 | | Crystal Structure of Nitrophorin 4 from Rhodnius Prolixus Complexed with Histamine at 1.5 A Resolution | | Descriptor: | HISTAMINE, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qiu, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
1IKF
 
 | | A CONFORMATION OF CYCLOSPORIN A IN AQUEOUS ENVIRONMENT REVEALED BY THE X-RAY STRUCTURE OF A CYCLOSPORIN-FAB COMPLEX | | Descriptor: | CYCLOSPORIN A, IGG1-KAPPA R45-45-11 FAB (HEAVY CHAIN), IGG1-KAPPA R45-45-11 FAB (LIGHT CHAIN) | | Authors: | Vix, O, Altschuh, D, Rees, B, Thierry, J.-C. | | Deposit date: | 1993-12-09 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformation of Cyclosporin a in Aqueous Environment Revealed by the X-Ray Structure of a Cyclosporin-Fab Complex.
Science, 256, 1992
|
|
1IKG
 
 | | MICHAELIS COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
1IKI
 
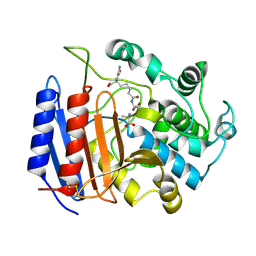 | | COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH THE PRODUCTS OF A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANINE, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
1IKJ
 
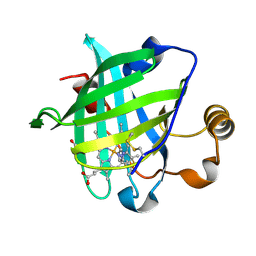 | | 1.27 A CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS COMPLEXED WITH IMIDAZOLE | | Descriptor: | IMIDAZOLE, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qui, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
1IKK
 
 | |
1IKL
 
 | |
1IKM
 
 | |
1IKN
 
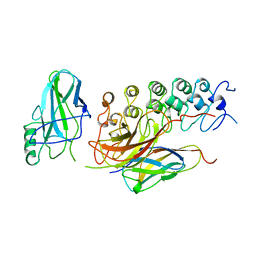 | | IKAPPABALPHA/NF-KAPPAB COMPLEX | | Descriptor: | PROTEIN (I-KAPPA-B-ALPHA), PROTEIN (NF-KAPPA-B P50D SUBUNIT), PROTEIN (NF-KAPPA-B P65 SUBUNIT) | | Authors: | Huxford, T, Huang, D.-B, Malek, S, Ghosh, G. | | Deposit date: | 1998-11-13 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation.
Cell(Cambridge,Mass.), 95, 1998
|
|
1IKO
 
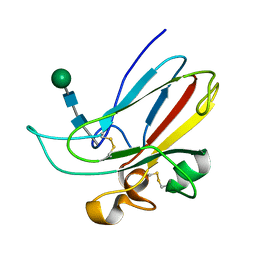 | | CRYSTAL STRUCTURE OF THE MURINE EPHRIN-B2 ECTODOMAIN | | Descriptor: | EPHRIN-B2, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Toth, J, Cutforth, T, Gelinas, A.D, Bethoney, K.A, Bard, J, Harrison, C.J. | | Deposit date: | 2001-05-03 | | Release date: | 2002-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of an ephrin ectodomain.
Dev.Cell, 1, 2001
|
|
1IKP
 
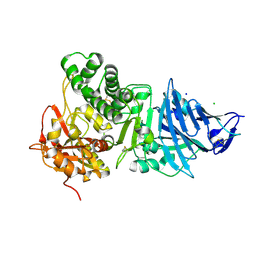 | | Pseudomonas Aeruginosa Exotoxin A, P201Q, W281A mutant | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
1IKQ
 
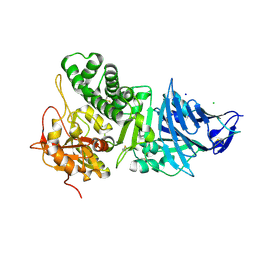 | | Pseudomonas Aeruginosa Exotoxin A, wild type | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
1IKT
 
 | |
1IKU
 
 | | myristoylated recoverin in the calcium-free state, NMR, 22 structures | | Descriptor: | MYRISTIC ACID, RECOVERIN | | Authors: | Tanaka, T, Ames, J.B, Harvey, T.S, Stryer, L, Ikura, M. | | Deposit date: | 1996-01-18 | | Release date: | 1996-07-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state.
Nature, 376, 1995
|
|
1IKV
 
 | | K103N Mutant HIV-1 Reverse Transcriptase in Complex with Efivarenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1IKW
 
 | | Wild Type HIV-1 Reverse Transcriptase in Complex with Efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1IKX
 
 | |
1IKY
 
 | | HIV-1 Reverse Transcriptase in Complex with the Inhibitor MSC194 | | Descriptor: | 1-[2-(3-ACETYL-2-HYDROXY-6-METHOXY-PHENYL)-CYCLOPROPYL]-3-(5-CYANO-PYRIDIN-2-YL)-THIOUREA, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
