4V10
 
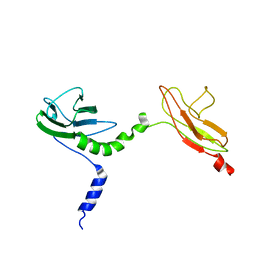 | | Skelemin Association with alpha2b,beta3 Integrin: A Structural Model | | Descriptor: | MYOMESIN-1 | | Authors: | Gorbatyuk, V, Deshmukh, L, Nguyen, K, Vinogradova, O. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Skelemin Association with Alphaiibbeta3 Integrin: A Structural Model.
Biochemistry, 53, 2014
|
|
5MO9
 
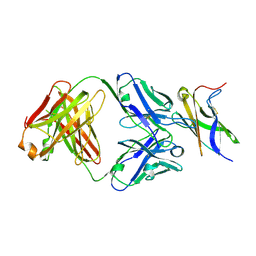 | |
5AEA
 
 | |
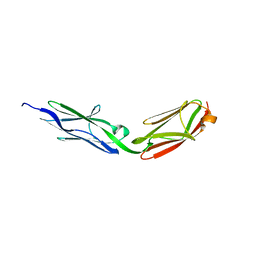
3LCY
 
 | |
6MG9
 
 | |
6SDB
 
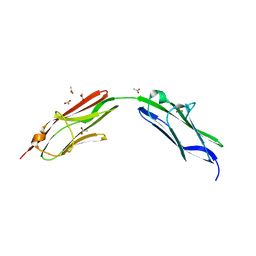 | | Chimeric titin Z1Z2 functionalized with a KLER exogenous peptide from decorin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Fleming, J.R, Hill, C, Mayans, O.M. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The ZT Biopolymer: A Self-Assembling Protein Scaffold for Stem Cell Applications.
Int J Mol Sci, 20, 2019
|
|
7AHS
 
 | | titin-N2A Ig81-Ig83 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Isoform 5 of Titin, ... | | Authors: | Fleming, J.R, Mayans, O. | | Deposit date: | 2020-09-25 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The N2A region of titin has a unique structural configuration.
J.Gen.Physiol., 153, 2021
|
|
6C6M
 
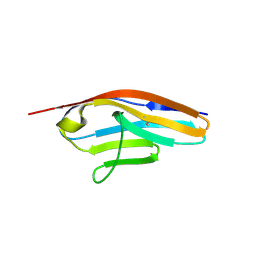 | | IgCam3 of human MLCK1 | | Descriptor: | Myosin light chain kinase, smooth muscle | | Authors: | Zuccola, H.J, Turner, J. | | Deposit date: | 2018-01-19 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intracellular MLCK1 diversion reverses barrier loss to restore mucosal homeostasis.
Nat. Med., 25, 2019
|
|
6YJ0
 
 | |
1NCT
 
 | | TITIN MODULE M5, N-TERMINALLY EXTENDED, NMR | | Descriptor: | TITIN | | Authors: | Pfuhl, M, Pastore, A. | | Deposit date: | 1996-08-13 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | When a module is also a domain: the role of the N terminus in the stability and the dynamics of immunoglobulin domains from titin.
J.Mol.Biol., 265, 1997
|
|
4UOW
 
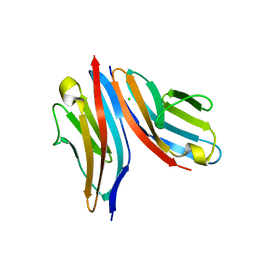 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | CHLORIDE ION, Obscurin, SODIUM ION, ... | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
4V2A
 
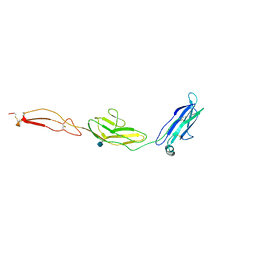 | | human Unc5A ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NETRIN RECEPTOR UNC5A | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
7R68
 
 | | Human obscurin Ig12 | | Descriptor: | Obscurin | | Authors: | Mauriello, G.E, Wright, N.T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminus of obscurin is flexible in solution.
Proteins, 91, 2023
|
|
7R67
 
 | | Human obscurin Ig13 | | Descriptor: | Obscurin | | Authors: | Moncure, G.E, Wright, N.T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminus of obscurin is flexible in solution.
Proteins, 91, 2023
|
|
4CBP
 
 | | Crystal structure of neural ectodermal development factor IMP-L2. | | Descriptor: | GLYCEROL, NEURAL/ECTODERMAL DEVELOPMENT FACTOR IMP-L2 | | Authors: | Kulahin, N, Kristensen, O, Brzozowski, M, Schluckebier, G, Meyts, P.D. | | Deposit date: | 2013-10-15 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Analysis of Imp-L2 Function
To be Published
|
|
3Q5O
 
 | |
4C4K
 
 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | 1,2-ETHANEDIOL, OBSCURIN, TITIN | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2013-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
3QP3
 
 | |
7LU6
 
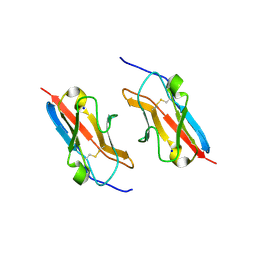 | | Crystal structure of the mouse Kirrel3 D1 homodimer | | Descriptor: | Kin of IRRE-like protein 3, SODIUM ION | | Authors: | Roman, C.A, Pak, J.S, Wang, J, Ozkan, E. | | Deposit date: | 2021-02-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep, 37, 2021
|
|
3RBS
 
 | |
7SJL
 
 | |
6NRX
 
 | | Crystal structure of DIP-eta IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dpr-interacting protein eta, isoform B, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
4QEG
 
 | |
6NRQ
 
 | | Crystal structure of Dpr10 IG1 bound to DIP-alpha IG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Defective proboscis extension response 10, isoform A, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
