8G0M
 
 | | Structure of complex between TV6.6 and CD98hc ECD | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, TETRAETHYLENE GLYCOL, ... | | Authors: | Kariolis, M.S, Lexa, K, Liau, N.P.D, Srivastava, D, Tran, H, Wells, R.C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD98hc is a target for brain delivery of biotherapeutics.
Nat Commun, 14, 2023
|
|
7XDQ
 
 | | Crystal structure of a glucosylglycerol phosphorylase mutant from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase, LITHIUM ION, beta-D-glucopyranose | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
7XDR
 
 | | Crystal structure of a glucosylglycerol phosphorylase from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
4WLC
 
 | | Structure of dextran glucosidase with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,6-alpha-glucosidase, ... | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
7QQJ
 
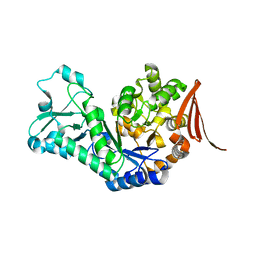 | | Sucrose phosphorylase from Jeotgalibaca ciconiae | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Sucrose phosphorylase | | Authors: | Ubiparip, Z, Capra, N, Rozeboom, H.J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2022-01-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sucrose phosphorylase from Jeotgalibaca ciconiae
To Be Published
|
|
7QQI
 
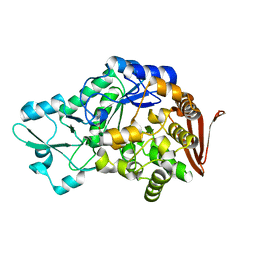 | | Sucrose phosphorylase from Faecalibaculum rodentium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aamy domain-containing protein | | Authors: | Ubiparip, Z, Capra, N, Rozeboom, H.J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2022-01-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Sucrose phosphorylase from Faecalibaculum rodentium
To Be Published
|
|
8SLV
 
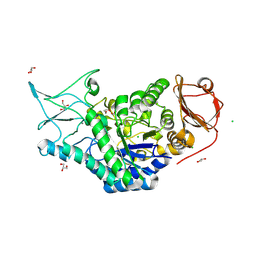 | | Structure of a salivary alpha-glucosidase from the mosquito vector Aedes aegypti. | | Descriptor: | 1,3-PROPANDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gittis, A.G, Williams, A.E, Garboczi, D, Calvo, E. | | Deposit date: | 2023-04-24 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and functional comparisons of salivary alpha-glucosidases from the mosquito vectors Aedes aegypti, Anopheles gambiae, and Culex quinquefasciatus.
Insect Biochem.Mol.Biol., 167, 2024
|
|
8IM8
 
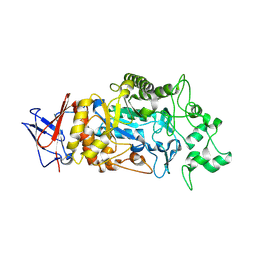 | | Crystal structure of Periplasmic alpha-amylase (MalS) from E.coli | | Descriptor: | CALCIUM ION, Periplasmic alpha-amylase | | Authors: | An, Y, Park, J.T, Park, K.H, Woo, E.J. | | Deposit date: | 2023-03-06 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Distinctive Permutated Domain Structure of Periplasmic alpha-Amylase (MalS) from Glycoside Hydrolase Family 13 Subfamily 19.
Molecules, 28, 2023
|
|
4XB3
 
 | | Structure of dextran glucosidase | | Descriptor: | CALCIUM ION, Glucan 1,6-alpha-glucosidase, HEXAETHYLENE GLYCOL | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-12-16 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
8JNX
 
 | | alkaline amylase Amy703 with truncated of N-terminus domain | | Descriptor: | Alpha-amylase, CALCIUM ION | | Authors: | Xiang, L, Zhang, G, Zhou, J. | | Deposit date: | 2023-06-06 | | Release date: | 2023-12-13 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.20279884 Å) | | Cite: | N-terminal domain truncation yielded a unique dimer of polysaccharide hydrolase with enhanced enzymatic activity, stability and calcium ion independence.
Int.J.Biol.Macromol., 266, 2024
|
|
7NF7
 
 | | Ovine rBAT ectodomain homodimer, asymmetric unit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lee, Y, Kuehlbrandt, W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Ca 2+ -mediated higher-order assembly of heterodimers in amino acid transport system b 0,+ biogenesis and cystinuria.
Nat Commun, 13, 2022
|
|
7JJT
 
 | | Ruminococcus bromii amylase Amy5 (RBR_07800) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alpha-amylase, ... | | Authors: | Cerqueira, F, Koropatkin, N. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The structures of the GH13_36 amylases from Eubacterium rectale and Ruminococcus bromii reveal subsite architectures that favor maltose production
Amylase, 4, 2020
|
|
5M99
 
 | | Functional Characterization and Crystal Structure of Thermostable Amylase from Thermotoga petrophila, reveals High Thermostability and an Archaic form of Dimerization | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-amylase, ... | | Authors: | Hameed, U, Price, I, Mirza, O.A. | | Deposit date: | 2016-11-01 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional characterization and crystal structure of thermostable amylase from Thermotoga petrophila, reveals high thermostability and an unusual form of dimerization.
Biochim. Biophys. Acta, 1865, 2017
|
|
6LGE
 
 | | Bombyx mori GH13 sucrose hydrolase complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6LGF
 
 | | Bombyx mori GH13 sucrose hydrolase mutant D247N complexed with sucrose | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Sucrose hydrolase, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6LGB
 
 | | Bombyx mori GH13 sucrose hydrolase complexed with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6S9V
 
 | | Crystal structure of sucrose 6F-phosphate phosphorylase from Thermoanaerobacter thermosaccharolyticum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Capra, N, Franceus, J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Comparison of a Promiscuous and a Highly Specific Sucrose 6 F -Phosphate Phosphorylase.
Int J Mol Sci, 20, 2019
|
|
6LGD
 
 | | Bombyx mori GH13 sucrose hydrolase complexed with 1,4-dideoxy-1,4-imino-D-arabinitol | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-D-ARABINITOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6LCV
 
 | |
6LGH
 
 | | Bombyx mori GH13 sucrose hydrolase mutant E322Q covalent intermediate | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Sucrose hydrolase, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6LGA
 
 | | Bombyx mori GH13 sucrose hydrolase | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6LGG
 
 | | Bombyx mori GH13 sucrose hydrolase mutant E322Q complexed with sucrose | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Sucrose hydrolase, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6LGI
 
 | |
6LGC
 
 | | Bombyx mori GH13 sucrose hydrolase complexed with 1-deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, GLYCEROL, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6LCU
 
 | |
