4UQJ
 
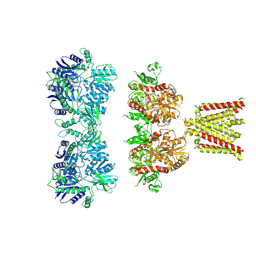 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
8HDK
 
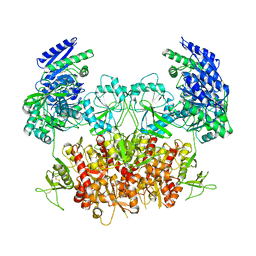 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (minor class in symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4UQK
 
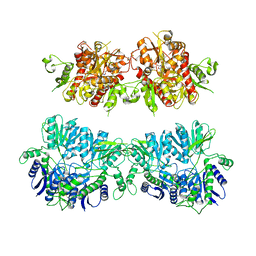 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
6PEQ
 
 | |
3C32
 
 | |
4E0W
 
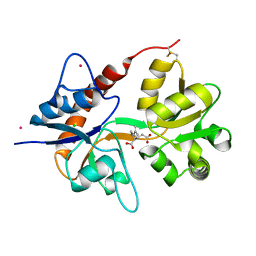 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | Kainate induces various domain closures in AMPA and kainate receptors.
Neurochem Int, 61, 2012
|
|
1GR2
 
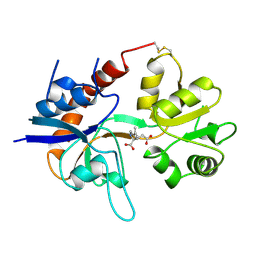 | | STRUCTURE OF A GLUTAMATE RECEPTOR LIGAND BINDING CORE (GLUR2) COMPLEXED WITH KAINATE | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, PROTEIN (GLUTAMATE RECEPTOR 2) | | Authors: | Armstrong, N, Sun, Y, Chen, G.Q, Gouaux, E. | | Deposit date: | 1998-09-17 | | Release date: | 1998-12-09 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a glutamate-receptor ligand-binding core in complex with kainate.
Nature, 395, 1998
|
|
5BUU
 
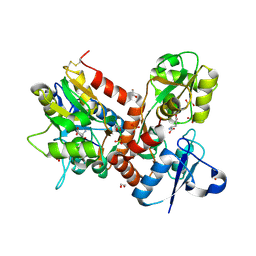 | | Crystal structure of the GluA2 ligand-binding domain (L483Y-N754S) in complex with glutamate and BPAM-321 at 2.07 A resolution | | Descriptor: | (3R)-7-chloro-2,3,4-trimethyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, 1,2-ETHANEDIOL, GLUTAMIC ACID, ... | | Authors: | Larsen, A.P, Tapken, D, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis and Pharmacology of Mono-, Di-, and Trialkyl-Substituted 7-Chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides Combined with X-ray Structure Analysis to Understand the Unexpected Structure-Activity Relationship at AMPA Receptors.
Acs Chem Neurosci, 7, 2016
|
|
5U8C
 
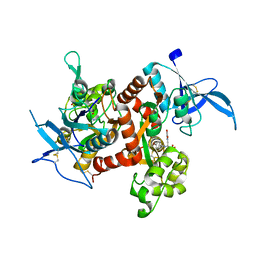 | |
4X48
 
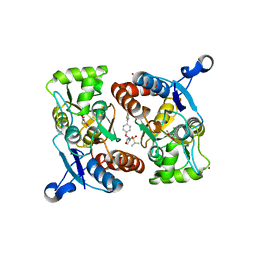 | | Crystal structure of GluR2 ligand-binding core | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, N-{(3S,4S)-4-[4-(5-cyanothiophen-2-yl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide, ... | | Authors: | Pandit, J. | | Deposit date: | 2014-12-02 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Discovery and Characterization of the alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptor Potentiator N-{(3S,4S)-4-[4-(5-Cyano-2-thienyl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide (PF-04958242).
J.Med.Chem., 58, 2015
|
|
4WXJ
 
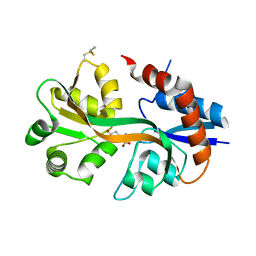 | | Drosophila muscle GluRIIB complex with glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor IIB,Glutamate receptor IIB | | Authors: | Dharkar, P, Mayer, M.L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Functional reconstitution of Drosophila melanogaster NMJ glutamate receptors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4MF3
 
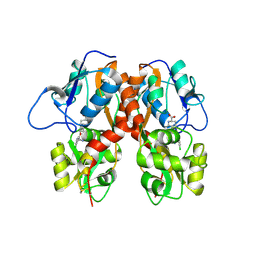 | | Crystal Structure of Human GRIK1 complexed with a 6-(tetrazolyl)aryl decahydroisoquinoline antagonist | | Descriptor: | (3S,4aS,6S,8aR)-6-[3-chloro-2-(1H-tetrazol-5-yl)phenoxy]decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 1 | | Authors: | Martinez-Perez, J.A, Iyengar, S, Shannon, H.E, Bleakman, D, Alt, A, Clawson, D.K, Arnold, B.M, Bell, M.G, Bleisch, T.J, Castano, A.M, Del Prado, M, Dominguez, E, Escribano, A.M, Filla, S.A, Ho, K.H, Hudziak, K.J, Jones, C.K, Katofiasc, M.A, Mateo, A, Mathes, B.M, Mattiuz, E.L, Ogden, A.M.L, Phebus, L.A, Simmons, R.M.A, Stack, D.R, Stratford, R.E, Winter, M.A, Wu, Z, Ornstein, P.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GluK1 antagonists from 6-(tetrazolyl)phenyl decahydroisoquinoline derivatives: in vitro profile and in vivo analgesic efficacy.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LZ7
 
 | |
4MH5
 
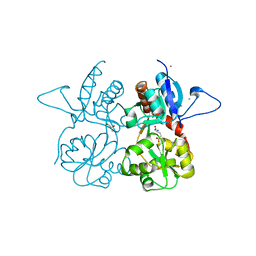 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
2OJT
 
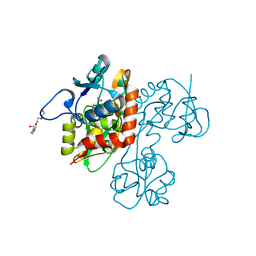 | | Structure and mechanism of kainate receptor modulation by anions | | Descriptor: | (S)-1-(2-AMINO-2-CARBOXYETHYL)-3(2-CARBOXYTHIOPHENE-3-YL-METHYL)-5-METHYLPYRIMIDINE-2,4-DIONE, BROMIDE ION, Glutamate receptor, ... | | Authors: | Mayer, M.L. | | Deposit date: | 2007-01-14 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and mechanism of kainate receptor modulation by anions.
Neuron, 53, 2007
|
|
2PBW
 
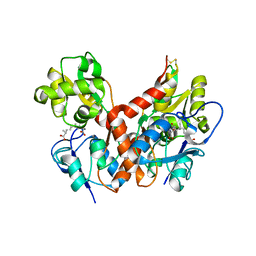 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex with the Partial agonist Domoic Acid at 2.5 A Resolution | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, Glutamate receptor, ionotropic kainate 1 | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
2P2A
 
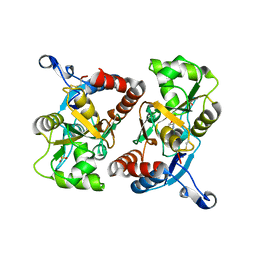 | | X-ray structure of the GluR2 ligand binding core (S1S2J) in complex with 2-Bn-tet-AMPA at 2.26A resolution | | Descriptor: | 2-AMINO-3-[3-HYDROXY-5-(2-BENZYL-2H-5-TETRAZOLYL)-4-ISOXAZOLYL]-PROPIONIC ACID, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Frydenvang, K, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A tetrazolyl-substituted subtype-selective AMPA receptor agonist.
J.Med.Chem., 50, 2007
|
|
1IIT
 
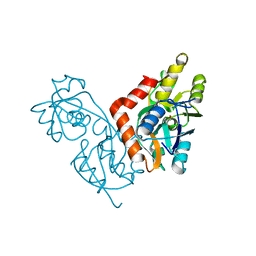 | | GLUR0 LIGAND BINDING CORE COMPLEX WITH L-SERINE | | Descriptor: | SERINE, Slr1257 protein | | Authors: | Mayer, M.L, Olson, R, Gouaux, E. | | Deposit date: | 2001-04-24 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state.
J.Mol.Biol., 311, 2001
|
|
1IIW
 
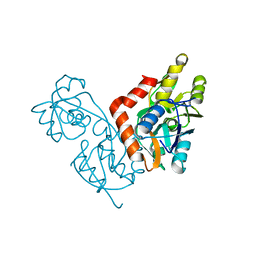 | |
1II5
 
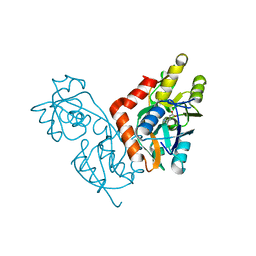 | |
1LB8
 
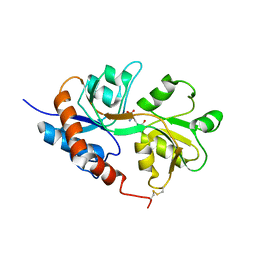 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with AMPA at 2.3 resolution | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
7OCA
 
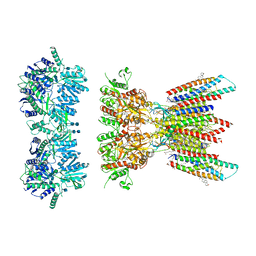 | | Resting state full-length GluA1/A2 heterotertramer in complex with TARP gamma 8 and CNIH2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, D, Watson, J.F, Matthews, P.M, Cais, O, Greger, I.H. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-09 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Gating and modulation of a hetero-octameric AMPA glutamate receptor.
Nature, 594, 2021
|
|
7OCC
 
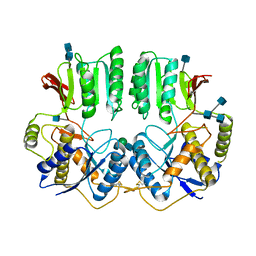 | | NTD of resting state GluA1/A2 heterotertramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 1, ... | | Authors: | Zhang, D, Watson, J.F, Matthews, P.M, Cais, O, Greger, I.H. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-09 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Gating and modulation of a hetero-octameric AMPA glutamate receptor.
Nature, 594, 2021
|
|
7OCD
 
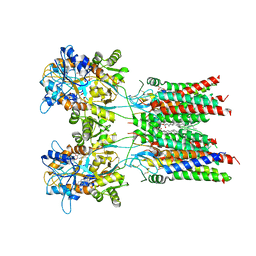 | | Resting state GluA1/A2 heterotetramer in complex with auxiliary subunit TARP gamma 8 (LBD-TMD) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, Glutamate receptor 2, ... | | Authors: | Zhang, D, Watson, J.F, Matthews, P.M, Cais, O, Greger, I.H. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-09 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Gating and modulation of a hetero-octameric AMPA glutamate receptor.
Nature, 594, 2021
|
|
7OCE
 
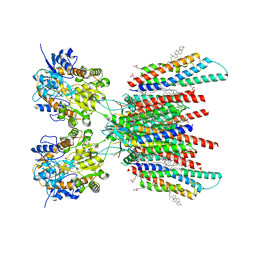 | | Resting state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and CNIH2 (LBD-TMD) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, CHOLESTEROL, ... | | Authors: | Zhang, D, Watson, J.F, Matthews, P.M, Cais, O, Greger, I.H. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-09 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Gating and modulation of a hetero-octameric AMPA glutamate receptor.
Nature, 594, 2021
|
|
