8E17
 
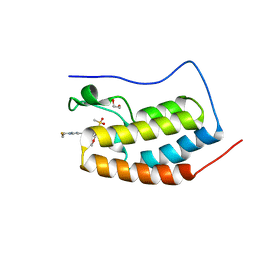 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P,6M)-6-[1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl]-4-[5-(methanesulfonyl)-2-methoxyphenyl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-09 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
6UJD
 
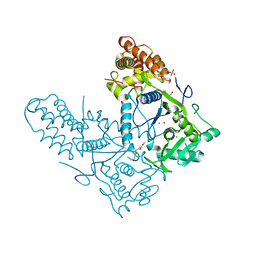 | |
6P4X
 
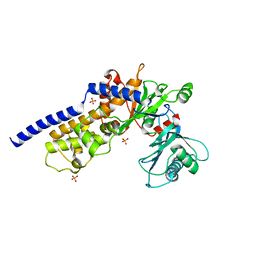 | |
6LYS
 
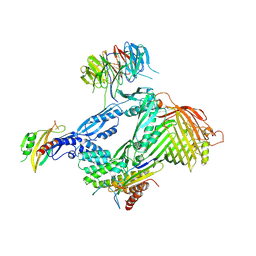 | | Structure of the BAM complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Xiao, L, Huang, Y. | | Deposit date: | 2020-02-15 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates.
Faseb J., 35, 2021
|
|
7MIU
 
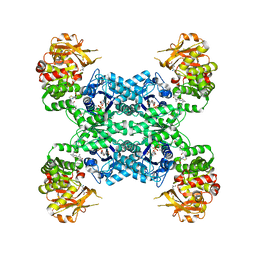 | | Mouse CTPS2 bound to inhibitor R80 | | Descriptor: | CTP synthase 2, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5PZA
 
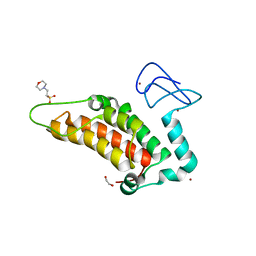 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 106) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
8E3W
 
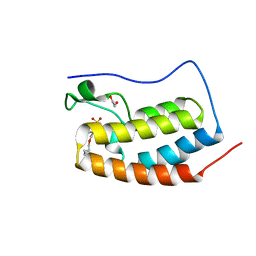 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P)-4-[2-(cyclopropylmethoxy)-5-(methanesulfonyl)phenyl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
7MN6
 
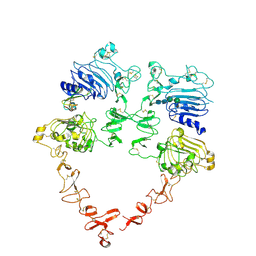 | | Structure of the HER2 S310F/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
6M01
 
 | | The structure of HitB-HitD complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Miyanaga, A, Kurihara, S, Kudo, F, Eguchi, T. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Complex of Adenylation Domain and Carrier Protein by Using Pantetheine Cross-Linking Probe.
Acs Chem.Biol., 15, 2020
|
|
6UJN
 
 | | P-glycoprotein mutant-C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
6UK0
 
 | | HIV-1 M184V reverse transcriptase-DNA complex | | Descriptor: | MAGNESIUM ION, Primer DNA, SULFATE ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-03 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75695229 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
6PC1
 
 | | Crystal structure of Helicobacter pylori PPX/GppA (E143A) in complex with ppGpp | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
7MSJ
 
 | | The crystal structure of mouse HVEM | | Descriptor: | SULFATE ION, Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
6M0J
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | Deposit date: | 2020-02-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
6M18
 
 | | ACE2-B0AT1 complex | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
8DYR
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P,6P)-4-[2-(cyclopropylmethoxy)-5-(methanesulfonyl)phenyl]-6-[1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-04 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
7A9I
 
 | |
6PCS
 
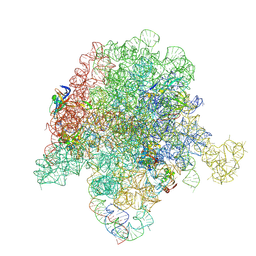 | | E. coli 50S ribosome bound to compound 40e | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl [4-(trifluoromethyl)phenyl]carbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6UKC
 
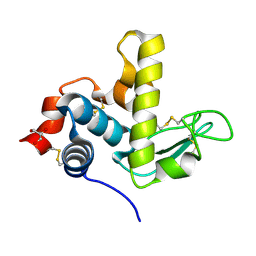 | |
6M2A
 
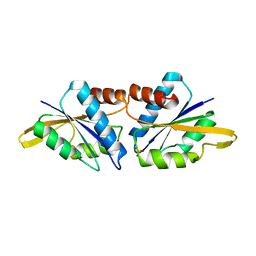 | |
7MSG
 
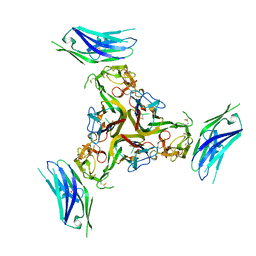 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
6P66
 
 | |
6UKY
 
 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 12 | | Descriptor: | 4-(6-{3-[2-(3-carboxypropanoyl)-6-methoxy-1-benzothiophen-4-yl]propyl}-5-methoxy-1-benzothiophen-2-yl)-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
8E7N
 
 | | Crystal structure of beluga whale Gammacoronavirus SW1 Mpro with GC-376 captured in two conformational states | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, GLYCEROL, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Inhibitor-Bound Main Protease from Delta- and Gamma-Coronaviruses.
Viruses, 15, 2023
|
|
8DK6
 
 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
