7UE7
 
 | | PANK3 complex structure with compound PZ-3883 | | Descriptor: | 1,2-ETHANEDIOL, 6-{4-[(4-cyclopropyl-3-fluorophenyl)acetyl]piperazin-1-yl}pyridazine-3-carbonitrile, ACETATE ION, ... | | Authors: | White, S.W, Yun, M, Lee, R.E. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of Brain Penetrant Pyridazine Pantothenate Kinase Activators.
J.Med.Chem., 67, 2024
|
|
7UEU
 
 | |
7B3X
 
 | | Notum complex with ARUK3003748 | | Descriptor: | 6-(m-tolylthio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Fish, P, Jones, E.Y. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
141D
 
 | |
7E6E
 
 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in D-cycloserine-inhibition | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6F
 
 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in L-cycloserine-inhibition | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
9BVB
 
 | | Crystal structure of human CYP3A4 in complex with SJYHJ-075 | | Descriptor: | 2-(2-chlorophenyl)-N-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]acetamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
6E1M
 
 | | Structure of AtTPC1(DDE) reconstituted in saposin A | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CALCIUM ION, PALMITIC ACID, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
9BV7
 
 | | Crystal structure of human CYP3A4 in complex with SJ000310315 | | Descriptor: | 3-(2-chlorophenyl)-N-{[(2S,7M)-7-(pyridin-3-yl)-2,3-dihydro-1-benzofuran-2-yl]methyl}propanamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
7B8X
 
 | | Notum-Fragment 210 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
9BVC
 
 | | Crystal structure of human CYP3A4 in complex with SJYHJ-110 | | Descriptor: | Cytochrome P450 3A4, N-(2-chlorophenyl)-N-[(3-hydroxyphenyl)methyl]-N'-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]urea, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
5EST
 
 | | Crystallographic analysis of the inhibition of porcine pancreatic elastase by a peptidyl boronic acid: structure of a reaction intermediate | | Descriptor: | CALCIUM ION, ELASTASE, N~2~-[(benzyloxy)carbonyl]-N-[(1R,2S)-1-(dihydroxyboranyl)-2-methylbutyl]-L-alaninamide, ... | | Authors: | Takahashi, L.H, Radhakrishnan, R, Rosenfieldjunior, R.E, Meyerjunior, E.F. | | Deposit date: | 1989-05-15 | | Release date: | 1992-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystallographic analysis of the inhibition of porcine pancreatic elastase by a peptidyl boronic acid: structure of a reaction intermediate.
Biochemistry, 28, 1989
|
|
8A5I
 
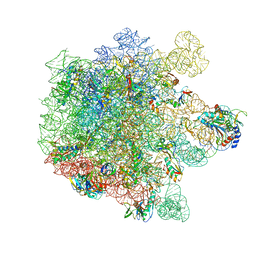 | | Cryo-EM structure of Lincomycin bound to the Listeria monocytogenes 50S ribosomal subunit. | | Descriptor: | 1,4-DIAMINOBUTANE, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for HflXr-mediated antibiotic resistance in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
6TEN
 
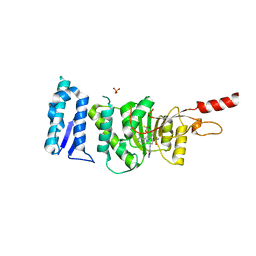 | | Crystal structure of Dot1L in complex with an inhibitor (compound 11). | | Descriptor: | 3-[(4-azanyl-6-methoxy-1,3,5-triazin-2-yl)amino]-4-[[(~{S})-[2,2-bis(fluoranyl)-1,3-benzodioxol-4-yl]-(3-chloranylpyridin-2-yl)methyl]amino]benzenesulfonamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
3KN5
 
 | |
6TFZ
 
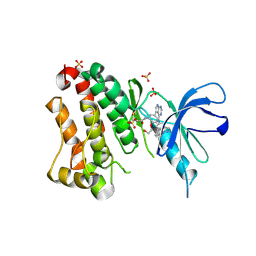 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 19 | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, SULFATE ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
3KRC
 
 | | Mint heterotetrameric geranyl pyrophosphate synthase in complex with IPP | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranyl diphosphate synthase large subunit, ... | | Authors: | Chang, T.-H, Hsieh, F.-L, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation
Plant Cell, 22, 2010
|
|
3KRP
 
 | | Mint heterotetrameric geranyl pyrophosphate synthase in complex with magnesium and GPP | | Descriptor: | 1,2-ETHANEDIOL, GERANYL DIPHOSPHATE, Geranyl diphosphate synthase large subunit, ... | | Authors: | Chang, T.-H, Hsieh, F.-L, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2009-11-19 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation
Plant Cell, 22, 2010
|
|
6AGR
 
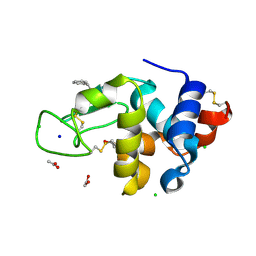 | | Structure of HEWL co-crystallised with phenylethyl alcohol | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYL-ETHANOL, ACETATE ION, ... | | Authors: | Seyedarabi, A, Seraj, Z. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The role of Cinnamaldehyde and Phenyl ethyl alcohol as two types of precipitants affecting protein hydration levels.
Int.J.Biol.Macromol., 146, 2019
|
|
6ELD
 
 | |
6JYZ
 
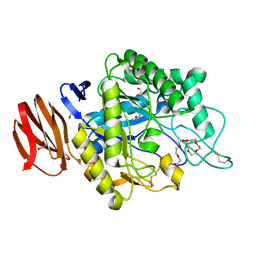 | | Crystal structure of endogalactoceramidase | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liuqing, C, Yan, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an endogalactosylceramidase from Rhodococcus hoagii 103S reveals the molecular basis of its substrate specificity.
J.Struct.Biol., 208, 2019
|
|
3L57
 
 | |
5X4M
 
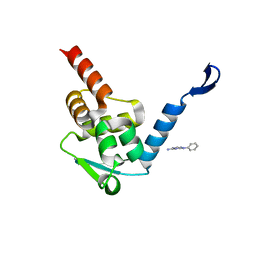 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1 | | Descriptor: | B-cell lymphoma 6 protein, N-phenyl-1,3,5-triazine-2,4-diamine | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
1JV7
 
 | | BACTERIORHODOPSIN O-LIKE INTERMEDIATE STATE OF THE D85S MUTANT AT 2.25 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
5FKR
 
 | | Unraveling the first step of xyloglucan degradation by the soil saprophyte Cellvibrio japonicus through the functional and structural characterization of a potent GH74 endo-xyloglucanase | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, ENDO-1,4-BETA-GLUCANASE/XYLOGLUCANASE, ... | | Authors: | Attia, M, Stepper, J, Davies, G.J, Brumer, H. | | Deposit date: | 2015-10-19 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Functional and Structural Characterization of a Potent Gh74 Endo-Xyloglucanase from the Soil Saprophyte Cellvibrio Japonicus Unravels the First Step of Xyloglucan Degradation.
FEBS J., 283, 2016
|
|
