4GF2
 
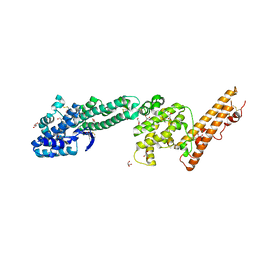 | | Crystal structure of Plasmodium falciparum Erythrocyte Binding Antigen 140 (PfEBA-140/BAEBL) | | Descriptor: | Erythrocyte binding antigen 140, GLYCEROL | | Authors: | Lin, D.H, Malpede, B.M, Batchelor, J.D, Tolia, N.H. | | Deposit date: | 2012-08-02 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal and Solution Structures of Plasmodium falciparum Erythrocyte-binding Antigen 140 Reveal Determinants of Receptor Specificity during Erythrocyte Invasion.
J.Biol.Chem., 287, 2012
|
|
8TRK
 
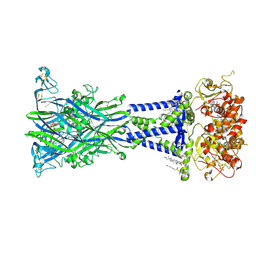 | | Cryo-EM structure of the rat P2X7 receptor in complex with the allosteric antagonist methyl blue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4',4''-[methanetriyltris(4,1-phenyleneazanediyl)]tri(benzene-1-sulfonic acid), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Oken, A.C, Ditter, I.A, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Mansoor, S.E. | | Deposit date: | 2023-08-09 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | P2X 7 receptors exhibit at least three modes of allosteric antagonism.
Sci Adv, 10, 2024
|
|
8TR8
 
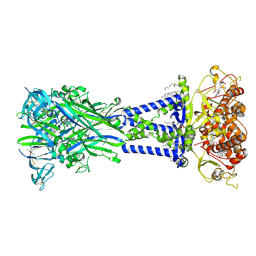 | | Cryo-EM structure of the rat P2X7 receptor in complex with the allosteric antagonist AZD9056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-DIPHOSPHATE, N-{[(3s,5s,7s)-adamantan-1-yl]methyl}-2-chloro-5-{3-[(3-hydroxypropyl)amino]propyl}benzamide, ... | | Authors: | Oken, A.C, Ditter, I.A, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Mansoor, S.E. | | Deposit date: | 2023-08-09 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | P2X 7 receptors exhibit at least three modes of allosteric antagonism.
Sci Adv, 10, 2024
|
|
8TRB
 
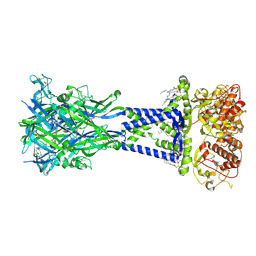 | | Cryo-EM structure of the rat P2X7 receptor in complex with the allosteric antagonist JNJ47965567 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-DIPHOSPHATE, N-{[4-(4-phenylpiperazin-1-yl)oxan-4-yl]methyl}-2-(phenylsulfanyl)pyridine-3-carboxamide, ... | | Authors: | Oken, A.C, Ditter, I.A, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Mansoor, S.E. | | Deposit date: | 2023-08-09 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | P2X 7 receptors exhibit at least three modes of allosteric antagonism.
Sci Adv, 10, 2024
|
|
8TRA
 
 | | Cryo-EM structure of the rat P2X7 receptor in complex with the allosteric antagonist GSK1482160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-DIPHOSPHATE, N-{[2-chloro-3-(trifluoromethyl)phenyl]methyl}-1-methyl-5-oxo-L-prolinamide, ... | | Authors: | Oken, A.C, Ditter, I.A, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Mansoor, S.E. | | Deposit date: | 2023-08-09 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | P2X 7 receptors exhibit at least three modes of allosteric antagonism.
Sci Adv, 10, 2024
|
|
8TR7
 
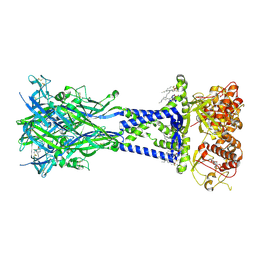 | | Cryo-EM structure of the rat P2X7 receptor in complex with the allosteric antagonist A839977 | | Descriptor: | (1P)-1-(2,3-dichlorophenyl)-N-({2-[(pyridin-2-yl)oxy]phenyl}methyl)-1H-tetrazol-5-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Oken, A.C, Ditter, I.A, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Mansoor, S.E. | | Deposit date: | 2023-08-09 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | P2X 7 receptors exhibit at least three modes of allosteric antagonism.
Sci Adv, 10, 2024
|
|
8TR6
 
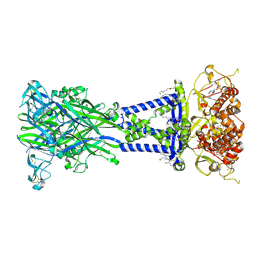 | | Cryo-EM structure of the rat P2X7 receptor in complex with the allosteric antagonist A438079 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-{[(5R)-5-(2,3-dichlorophenyl)tetrazolidin-1-yl]methyl}pyridine, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Oken, A.C, Ditter, I.A, Lisi, N.E, Krishnamurthy, I, McCarthy, A.E, Godsey, M.H, Mansoor, S.E. | | Deposit date: | 2023-08-09 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | P2X 7 receptors exhibit at least three modes of allosteric antagonism.
Sci Adv, 10, 2024
|
|
4NRA
 
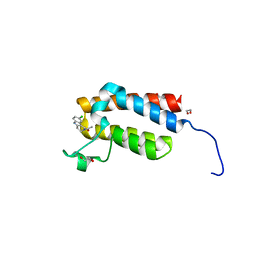 | | Crystal Structure of the bromodomain of human BAZ2B in complex with compound-6 E11322 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-chloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl)ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Ferguson, F.M, Filippakopoulos, P, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain.
J.Med.Chem., 56, 2013
|
|
7B4D
 
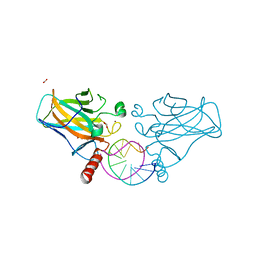 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R273C/S240R double mutant bound to DNA and MQ: R273C/S240R-DNA-MQ | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Diskin-Posner, Y, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
5IJX
 
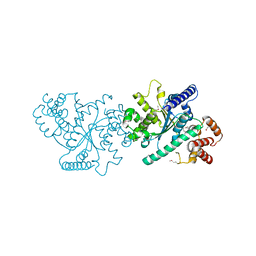 | |
2WOV
 
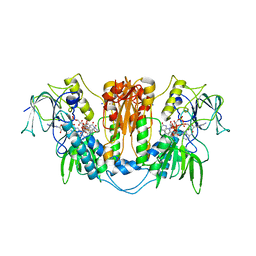 | | Trypanosoma brucei trypanothione reductase with bound NADP. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
3F8J
 
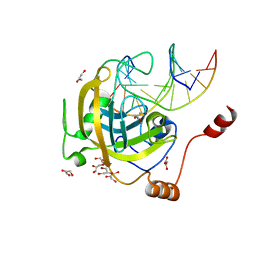 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group C222(1) | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1, ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
2WPF
 
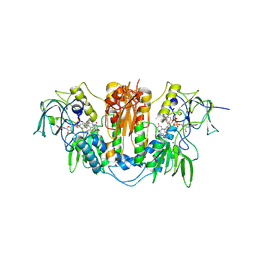 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00085762) | | Descriptor: | 3-[(4S)-6-CHLORO-2-METHYL-4-(4-METHYLPHENYL)QUINAZOLIN-3(4H)-YL]-N,N-DIMETHYLPROPAN-1-AMINE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WOW
 
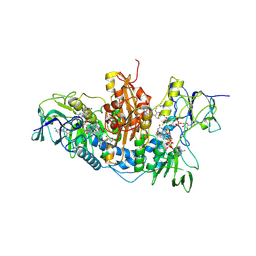 | | Trypanosoma brucei trypanothione reductase with NADP and trypanothione bound | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
8PPL
 
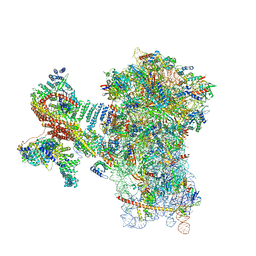 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
3C6N
 
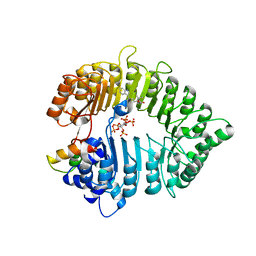 | | Small molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling | | Descriptor: | (2S)-8-[(tert-butoxycarbonyl)amino]-2-(1H-indol-3-yl)octanoic acid, INOSITOL HEXAKISPHOSPHATE, SKP1-like protein 1A, ... | | Authors: | Tan, X, Zheng, N, Hayashi, K. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-22 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4C33
 
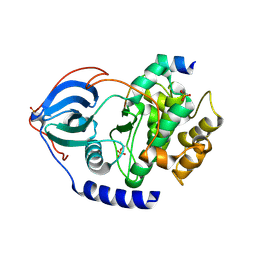 | | PKA-S6K1 Chimera Apo | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
9J7I
 
 | | Cryo-EM Structure of calcium sensing receptor in complex gamma-glutamyl-valyl-glycine as a kokumi substance | | Descriptor: | Extracellular calcium-sensing receptor, gamma-glutamyl-valyl-glycine | | Authors: | Yamaguchi, H, Kitajima, S, Suzuki, H, Suzuki, S, Nishikawa, K, Maruyama, Y, Kamegawa, A, Kazutoshi, T, Tagami, U, Kuroda, M, Fujiyoshi, Y, Sugiki, M. | | Deposit date: | 2024-08-19 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM structure of the calcium-sensing receptor complexed with the kokumi substance gamma-glutamyl-valyl-glycine.
Sci Rep, 15, 2025
|
|
6YCH
 
 | | Crystal structure of GcoA T296A bound to guaiacol | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Ellis, E.S, DuBois, J.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCK
 
 | | Crystal structure of GcoA T296A bound to p-vanillin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCO
 
 | | Crystal structure of GcoA F169S bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
7K98
 
 | | Preaminoacylation complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine (F-AMS) | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-28 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
2Z3J
 
 | | Crystal structure of blasticidin S deaminase (BSD) R90K mutant | | Descriptor: | Blasticidin-S deaminase, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Teh, A.H, Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
7K9M
 
 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
2BJ4
 
 | | ESTROGEN RECEPTOR ALPHA LBD IN COMPLEX WITH A PHAGE-DISPLAY DERIVED PEPTIDE ANTAGONIST | | Descriptor: | 4-HYDROXYTAMOXIFEN, ESTROGEN RECEPTOR, PEPTIDE ANTAGONIST | | Authors: | Kong, E, Heldring, N, Gustafsson, J.A, Treuter, E, Hubbard, R.E, Pike, A.C.W. | | Deposit date: | 2005-01-28 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Delineation of a Unique Protein-Protein Interaction Site on the Surface of the Estrogen Receptor
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
