4Z40
 
 | | Active site complex BamBC of Benzoyl Coenzyme A reductase as isolated | | Descriptor: | Benzoyl-CoA reductase, putative, IRON/SULFUR CLUSTER, ... | | Authors: | Weinert, T, Kung, J.W, Weidenweber, S, Huwiler, S.G, Boll, M, Ermler, U. | | Deposit date: | 2015-04-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of enzymatic benzene ring reduction.
Nat.Chem.Biol., 11, 2015
|
|
3LGN
 
 | | Crystal structure of IsdI in complex with heme | | Descriptor: | Heme-degrading monooxygenase isdI, MAGNESIUM ION, OXYGEN MOLECULE, ... | | Authors: | Ukpabi, G.N, Murphy, M.E.P. | | Deposit date: | 2010-01-20 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The IsdG-family of haem oxygenases degrades haem to a novel chromophore
Mol.Microbiol., 75, 2010
|
|
3E84
 
 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, Acylneuraminate cytidylyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
8KCZ
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione
To Be Published
|
|
3EL3
 
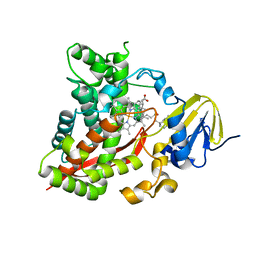 | | Distinct Monooxygenase and Farnesene Synthase Active Sites in Cytochrome P450 170A1 | | Descriptor: | (3S,3aR,6S)-3,7,7,8-tetramethyl-2,3,4,5,6,7-hexahydro-1H-3a,6-methanoazulene, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Zhao, B, Waterman, M.R. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site
J.Biol.Chem., 284, 2009
|
|
1BF2
 
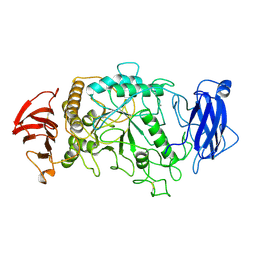 | | STRUCTURE OF PSEUDOMONAS ISOAMYLASE | | Descriptor: | CALCIUM ION, ISOAMYLASE | | Authors: | Katsuya, Y, Mezaki, Y, Kubota, M, Matsuura, Y. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Pseudomonas isoamylase at 2.2 A resolution.
J.Mol.Biol., 281, 1998
|
|
8I5Z
 
 | |
1B3Z
 
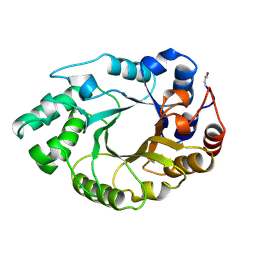 | | XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOPENTAOSE | | Descriptor: | PROTEIN (XYLANASE), beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Schmidt, A, Kratky, C. | | Deposit date: | 1998-12-15 | | Release date: | 1999-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Xylan binding subsite mapping in the xylanase from Penicillium simplicissimum using xylooligosaccharides as cryo-protectant.
Biochemistry, 38, 1999
|
|
8I0C
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S0703 | | Descriptor: | 1-[4-[3,5-bis(chloranyl)phenyl]-3-fluoranyl-phenyl]cyclopropane-1-carboxylic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, He, S, Liu, Y, Fang, P, Sun, H. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
9FRV
 
 | | Arginase 2 in complex with inhibitor | | Descriptor: | Arginase-2, mitochondrial, MANGANESE (II) ION, ... | | Authors: | Petersen, j. | | Deposit date: | 2024-06-19 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Design and Synthesis of Acyclic Boronic Acid Arginase Inhibitors.
J.Med.Chem., 67, 2024
|
|
3EBA
 
 | | CAbHul6 FGLW mutant (humanized) in complex with human lysozyme | | Descriptor: | CAbHul6, Lysozyme C, SULFATE ION | | Authors: | Loris, R, Vincke, C, Saerens, D, Martinez-Rodriguez, S, Muyldermans, S, Conrath, K. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | General Strategy to Humanize a Camelid Single-domain Antibody and Identification of a Universal Humanized Nanobody Scaffold
J.Biol.Chem., 284, 2009
|
|
8OFW
 
 | |
4Z82
 
 | | Cysteine bound rat cysteine dioxygenase C164S variant at pH 8.1 | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Fellner, M, Tchesnokov, E.P, Siakkou, E, Rutledge, M.T, Kanitz, M, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2015-04-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of cysteine 164 on active site structure in rat cysteine dioxygenase.
J.Biol.Inorg.Chem., 21, 2016
|
|
1B5U
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANT | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
4Z46
 
 | |
4Z85
 
 | |
9F7J
 
 | | Rhodococcus diDNase in apo state | | Descriptor: | diDNase | | Authors: | Mortensen, S, Sondermann, H. | | Deposit date: | 2024-05-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and bioinformatics analyses identify deoxydinucleotide-specific nucleases and their association with genomic islands in gram-positive bacteria.
Nucleic Acids Res., 53, 2025
|
|
3EEI
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from neisseria meningitidis in complex with methylthio-immucillin-A | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Ho, M, Rinaldo-matthis, A, Brown, R.L, Norris, G.E, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-09-04 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from neisseria meningitidis in complex with methylthio-immucillin-A
To be Published
|
|
1B9Z
 
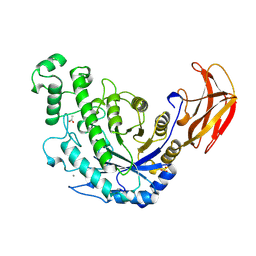 | | BACILLUS CEREUS BETA-AMYLASE COMPLEXED WITH MALTOSE | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
1BB5
 
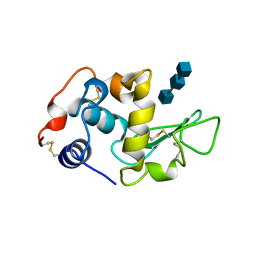 | |
9F7L
 
 | | Bartonella henselae NrnC bound to deoxy-pGG | | Descriptor: | 3'-5' exonuclease, CHLORIDE ION, DNA (5'-R(P*DGP*DG)-3'), ... | | Authors: | Mortensen, S, Sondermann, H. | | Deposit date: | 2024-05-04 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and bioinformatics analyses identify deoxydinucleotide-specific nucleases and their association with genomic islands in gram-positive bacteria.
Nucleic Acids Res., 53, 2025
|
|
9F7D
 
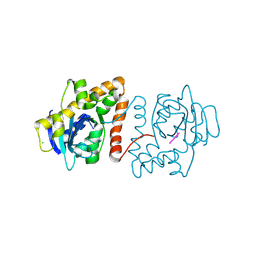 | | Rhodococcus diDNase bound to deoxy-pGG | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*GP*G)-3'), diDNase | | Authors: | Mortensen, S, Sondermann, H. | | Deposit date: | 2024-05-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and bioinformatics analyses identify deoxydinucleotide-specific nucleases and their association with genomic islands in gram-positive bacteria.
Nucleic Acids Res., 53, 2025
|
|
4Z9G
 
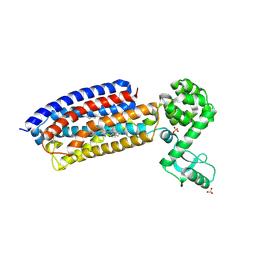 | | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 in a hexagonal setting with translational non-crystallographic symmetry | | Descriptor: | 3,6-dimethyl-N-(pentan-3-yl)-2-(2,4,6-trimethylphenoxy)pyridin-4-amine, Corticotropin-releasing factor receptor 1,Lysozyme,Corticotropin-releasing factor receptor 1, OLEIC ACID, ... | | Authors: | Dore, A.S, Bortolato, A, Hollenstein, K, Cheng, R.K.Y, Read, R.J, Marshall, F.H. | | Deposit date: | 2015-04-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.183 Å) | | Cite: | Decoding Corticotropin-Releasing Factor Receptor Type 1 Crystal Structures.
Curr Mol Pharmacol, 10, 2017
|
|
3LS9
 
 | | Crystal structure of atrazine chlorohydrolase TrzN from Arthrobacter aurescens TC1 complexed with zinc | | Descriptor: | Triazine hydrolase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Seffernick, J, Wackett, L.P, Almo, S.C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of atrazine chlorohydrolase TrzN
from Arthrobacter aurescens TC1 complexed with zinc
To be Published
|
|
9F7G
 
 | | Nocardioides diDNase bound to deoxy-pGG | | Descriptor: | DNA (5'-D(P*GP*G)-3'), Ribonuclease D, SULFATE ION | | Authors: | Mortensen, S, Sondermann, H. | | Deposit date: | 2024-05-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and bioinformatics analyses identify deoxydinucleotide-specific nucleases and their association with genomic islands in gram-positive bacteria.
Nucleic Acids Res., 53, 2025
|
|
