8UUE
 
 | |
8USX
 
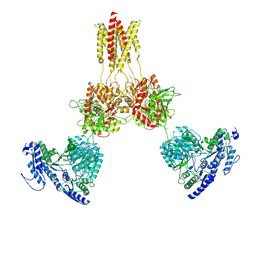 | | Glycine-bound GluN1a-3A NMDA receptor | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 3A | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
5HGQ
 
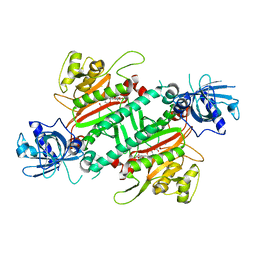 | | Loa loa Lysyl-tRNA synthetase in complex with Cladosporin. | | Descriptor: | GLYCEROL, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Sharma, A, Sharma, M, Yogavel, M, Sharma, A. | | Deposit date: | 2016-01-08 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Protein Translation Enzyme lysyl-tRNA Synthetase Presents a New Target for Drug Development against Causative Agents of Loiasis and Schistosomiasis
PLoS Negl Trop Dis, 10, 2016
|
|
8DEY
 
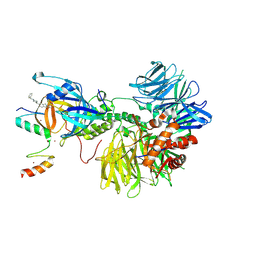 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
6PA5
 
 | |
3DIZ
 
 | |
6PA9
 
 | |
4V46
 
 | | Crystal structure of the BAFF-BAFF-R complex | | Descriptor: | MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Kim, H.M, Yu, K.S, Lee, M.E, Shin, D.R, Kim, Y.S, Paik, S.G, Yoo, O.J, Lee, H, Lee, J.-O. | | Deposit date: | 2003-03-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation
NAT.STRUCT.BIOL., 10, 2003
|
|
6GBW
 
 | | Thrombin in complex with MI2100 ((S)-N-(2-(aminomethyl)-5-chlorobenzyl)-1-((benzylsulfonyl)-L-arginyl)pyrrolidine-2-carboxamide) | | Descriptor: | (2~{S})-~{N}-[[2-(aminomethyl)-5-chloranyl-phenyl]methyl]-1-[(2~{S})-5-carbamimidamido-2-[(phenylmethyl)sulfonylamino]pentanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2018-04-16 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Strategies for Late-Stage Optimization: Profiling Thermodynamics by Preorganization and Salt Bridge Shielding.
J.Med.Chem., 62, 2019
|
|
8FTN
 
 | | E. coli ArnA dehydrogenase domain mutant - N492A | | Descriptor: | Bifunctional UDP-4-amino-4-deoxy-L-arabinose formyltransferase/UDP-glucuronic acid oxidase ArnA, SULFATE ION | | Authors: | Sousa, M.C, Mitchell, M.E. | | Deposit date: | 2023-01-12 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Conformational Change in ArnA Dehydrogenase for Selective Inhibition of Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
1MRG
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1RD4
 
 | | An allosteric inhibitor of LFA-1 bound to its I-domain | | Descriptor: | 1-ACETYL-4-(4-{4-[(2-ETHOXYPHENYL)THIO]-3-NITROPHENYL}PYRIDIN-2-YL)PIPERAZINE, Integrin alpha-L | | Authors: | Crump, M.P, Ceska, T.A, Spyracopoulos, L, Henry, A, Archibald, S.C, Alexander, R, Taylor, R.J, Findlow, S.C, O'Connell, J, Robinson, M.K, Shock, A. | | Deposit date: | 2003-11-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an allosteric inhibitor of LFA-1 bound to the I-domain studied by crystallography, NMR, and calorimetry
Biochemistry, 43, 2004
|
|
2WHE
 
 | | Structure of native Beta-Phosphoglucomutase in an open conformation without bound ligands. | | Descriptor: | BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic Details of Near-Transition State Conformers for Enzyme Phosphoryl Transfer Revealed by Mgf-3 Rather Than by Phosphoranes.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1WDN
 
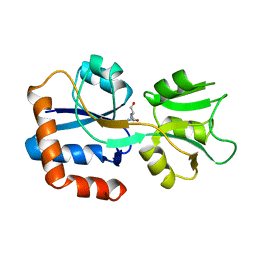 | | GLUTAMINE-BINDING PROTEIN | | Descriptor: | GLUTAMINE, GLUTAMINE BINDING PROTEIN | | Authors: | Sun, Y.-J, Rose, J, Wang, B.-C, Hsiao, C.-D. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of glutamine-binding protein complexed with glutamine at 1.94 A resolution: comparisons with other amino acid binding proteins.
J.Mol.Biol., 278, 1998
|
|
6PA2
 
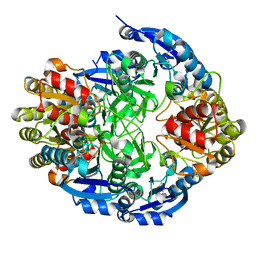 | |
4QY3
 
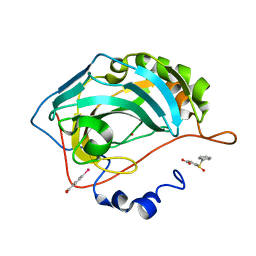 | |
3IC2
 
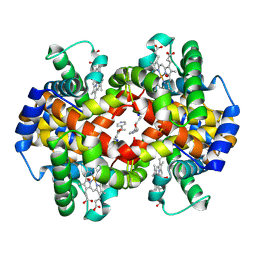 | | Crystal Structure of liganded hemoglobin in complex with a potent antisickling agent, INN-266 | | Descriptor: | 4-[(5-methoxy-2-methylphenoxy)methyl]pyridine, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Safo, M.K, Musayev, F.N, Gandhi, A.K, Jorge, P. | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and in Vitro Chracterization of Pyridyl Derivatives of Benzaldehydes: Highly Potent Antisickling Agents
To be Published
|
|
6PA4
 
 | |
3IIQ
 
 | |
4P73
 
 | | PheRS in complex with compound 1a | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6BQY
 
 | |
5ZRN
 
 | |
6YAA
 
 | | Structure of the (SR) Ca2+-ATPase bound to the inhibitor compound CAD204520 and TNP-ATP | | Descriptor: | 4-[2-[(2~{R})-2-[3-propyl-6-(trifluoromethyloxy)-1~{H}-indol-2-yl]piperidin-1-yl]ethyl]morpholine, POTASSIUM ION, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE, ... | | Authors: | Heit, S, Marchesini, M, Gherli, A, Montanaro, A, Patrizi, L, Sorrentino, C, Pagliaro, L, Rompietti, C, Kitara, S, Olesen, C.E, Moller, J.V, Savi, M, Bocchi, L, Vilella, R, Rizzi, F, Baglione, M, Rastelli, G, Loiacona, C, La Starza, R, Mecucci, C, Stegmair, K, Aversa, F, Stilli, D, Lund Winther, A.M, Sportoletti, P, Dalby-Brown, W, Roti, G, Bublitz, M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia.
Cell Chem Biol, 27, 2020
|
|
4NRB
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with compound-1 N01197 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, N-methyl-2-(tetrahydro-2H-pyran-4-yloxy)benzamide | | Authors: | Muniz, J.R.C, Felletar, I, Chaikuad, A, Filippakopoulos, P, Ferguson, F.M, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ciulli, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain.
J.Med.Chem., 56, 2013
|
|
