3ZH5
 
 | | The structure of Haemophilus influenzae protein E | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PROTEIN E | | Authors: | Singh, B, Al-Jubair, T, Riesbeck, K, Thunnissen, M.M.G.M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The Unique Structure of Haemophilus Influenzae Protein E Reveals Multiple Binding Sites for Host Factors.
Infect.Immun., 81, 2013
|
|
6RYJ
 
 | |
6S4K
 
 | | The crystal structure of glycogen phosphorylase in complex with 12 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(4-phenyl-1,3-thiazol-2-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
5ZRH
 
 | | M. smegmatis antimutator protein MutT2 in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-MONOPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5C4Y
 
 | | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, Putative transcription regulator Lmo0852 | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes
to be published
|
|
6G4Q
 
 | | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2 | | Descriptor: | 1,2-ETHANEDIOL, Succinate--CoA ligase [ADP-forming] subunit beta, mitochondrial, ... | | Authors: | Bailey, H.J, Shrestha, L, Rembeza, E, Sorrell, F.J, Newman, J, Strain-Damerell, C, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2
To Be Published
|
|
6RU7
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with double phosphorylated p63 PAD2P peptide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
7C4O
 
 | | Solution structure of the Orange domain from human protein HES1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Fan, J.S, Nayak, A, Swaminathan, K. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Induction of Transcriptional Inhibitor Hairy and Enhancer of Split Homolog-1 and the Related Repression of Tumor-Suppressor Thioredoxin-Interacting Protein Are Important Components of Cell-Transformation Program Imposed by Oncogenic Kinase Nucleophosmin-Anaplastic Lymphoma Kinase.
Am J Pathol, 2022
|
|
4YBG
 
 | | Crystal structure of the MAEL domain of Drosophila melanogaster Maelstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Protein maelstrom, ... | | Authors: | Matsumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystal Structure and Activity of the Endoribonuclease Domain of the piRNA Pathway Factor Maelstrom
Cell Rep, 11, 2015
|
|
6S1H
 
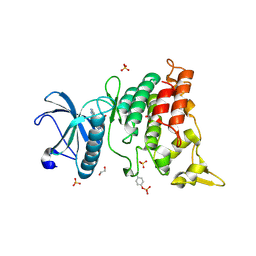 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
6S23
 
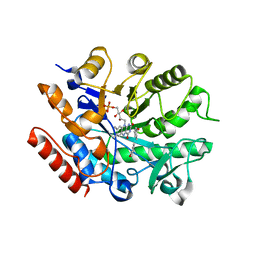 | | Crystal structure of ene-reductase GsOYE from Galleria sulphuraria in complex with 2-methyl-cyclopenten-1-one | | Descriptor: | 2-methylcyclopenta-2,4-dien-1-one, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Robescu, M.S, Niero, M, Hall, M, Bergantino, E, Cendron, L. | | Deposit date: | 2019-06-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Two new ene-reductases from photosynthetic extremophiles enlarge the panel of old yellow enzymes: CtOYE and GsOYE.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
6S4O
 
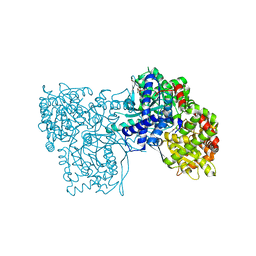 | | The crystal structure of glycogen phosphorylase in complex with 9 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-naphthalen-2-yl-1~{H}-imidazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
7D8U
 
 | | Crystal structure of the C-terminal domain of pNP868R from African swine fever virus | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GTP--RNA guanylyltransferase, ... | | Authors: | Du, X, Geng, Z, Zhang, H. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Biochemical Characteristic of the Methyltransferase (MTase) Domain of RNA Capping Enzyme from African Swine Fever Virus.
J.Virol., 2020
|
|
5AXV
 
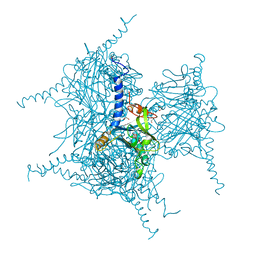 | | Crystal Structure of Cypovirus Polyhedra R13K Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Ijiri, H, Negishi, H, Yamanaka, H, Sasaki, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design of Enzyme-Encapsulated Protein Containers by In Vivo Crystal Engineering
Adv. Mater. Weinheim, 27, 2015
|
|
2V62
 
 | | Structure of vaccinia-related kinase 2 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SERINE/THREONINE-PROTEIN KINASE VRK2, ... | | Authors: | Bunkoczi, G, Eswaran, J, Cooper, C, Fedorov, O, Keates, T, Rellos, P, Salah, E, Savitsky, P, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Pseudokinase Vrk3 Reveals a Degraded Catalytic Site, a Highly Conserved Kinase Fold, and a Putative Regulatory Binding Site.
Structure, 17, 2009
|
|
6BBV
 
 | | Crystal Structure of JAK2 in complex with compound 25 | | Descriptor: | N-{cis-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Han, S. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of N-{cis-3-[Methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide (PF-04965842): A Selective JAK1 Clinical Candidate for the Treatment of Autoimmune Diseases.
J. Med. Chem., 61, 2018
|
|
7FXF
 
 | | Crystal Structure of human FABP4 in complex with 2-[(2-chlorophenoxy)methyl]-1,3-thiazole-4-carboxylic acid | | Descriptor: | 2-[(2-chlorophenoxy)methyl]-1,3-thiazole-4-carboxylic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5DNE
 
 | |
7FYS
 
 | | Crystal Structure of human FABP4 in complex with 7-(4-methoxyphenyl)-1,8-dithia-3-azaspiro[4.5]dec-6-ene-2,4-dione, i.e. SMILES [C@@]12(C=C(c3ccc(cc3)OC)SCC2)SC(=O)NC1=O with IC50=4.5 microM | | Descriptor: | (5R)-7-(4-hydroxyphenyl)-1,8-dithia-3-azaspiro[4.5]dec-6-ene-2,4-dione, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Widmer, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7G09
 
 | | Crystal Structure of human FABP4 in complex with 8-(3-bromophenyl)-7,9-dioxa-1-thia-3-azaspiro[4.5]decane-2,4-dione, i.e. SMILES [C@@]12(C(=O)NC(=O)S1)CO[C@@H](OC2)c1cc(ccc1)Br with IC50=2.3 microM | | Descriptor: | (5s,8s)-8-(3-bromophenyl)-7,9-dioxa-1-thia-3-azaspiro[4.5]decane-2,4-dione, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kumagai, Y, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7G0N
 
 | | Crystal Structure of delipidated apo human FABP4 | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FVW
 
 | | Crystal Structure of human FABP4 in complex with 2-[2-(3-chlorophenyl)ethyl]-1,3-thiazole-4-carboxylic acid | | Descriptor: | 2-[2-(3-chlorophenyl)ethyl]-1,3-thiazole-4-carboxylic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FYR
 
 | | Crystal Structure of human FABP4 in complex with 5-(3,4-dichlorophenyl)-5-methyl-2-sulfanylidene-1,3-diazinane-4,6-dione, i.e. SMILES C1(c2cc(c(cc2)Cl)Cl)(C(=O)NC(=S)NC1=O)C with IC50=3.3 microM | | Descriptor: | 5-(3,4-dichlorophenyl)-5-methyl-2-sulfanylidene-1,3-diazinane-4,6-dione, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Borgulya, J, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FX0
 
 | | Crystal Structure of human FABP4 in complex with 5-[(2-chloroanilino)methyl]-4-ethyl-1,2,4-triazole-3-thiol, i.e. SMILES N1(C(=NN=C1CNc1c(Cl)cccc1)S)CC with IC50=0.705 microM | | Descriptor: | 5-[(2-chloroanilino)methyl]-4-ethyl-4H-1,2,4-triazole-3-thiol, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FXD
 
 | | Crystal Structure of human FABP5 in complex with 2-(indole-1-carbonylamino)benzoic acid, i.e. SMILES c12N(C(=O)Nc3c(cccc3)C(=O)O)C=Cc1cccc2 with IC50=18.1696 microM | | Descriptor: | 2-[(2,3-dihydro-1H-indole-1-carbonyl)amino]benzoic acid, Fatty acid-binding protein 5 | | Authors: | Ehler, A, Benz, J, Obst, U, Ning, R, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
