7QOT
 
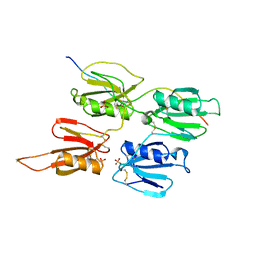 | | Factor XI and Plasma Kallikrein apple domain structures reveals different kininogen bound complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa heavy chain, Kininogen-1 light chain, ... | | Authors: | Li, C, Awital, B, Wong, S, Dreveny, I, Meijers, J, Emsley, J. | | Deposit date: | 2021-12-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J Thromb Haemost, 17, 2019
|
|
6JCV
 
 | | Cryo-EM structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) with Mg2+ at pH7.5 | | Descriptor: | MAGNESIUM ION, Putative ketol-acid reductoisomerase 2 | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6JCZ
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADPH, and CPD at pH7.5 | | Descriptor: | MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
4BHB
 
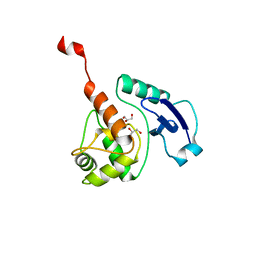 | | Crystal structure of Mycobacterium tuberculosis O6-METHYLGUANINE METHYLTRANSFERASE | | Descriptor: | GLYCEROL, METHYLATED-DNA--PROTEIN-CYSTEINE METHYLTRANSFERASE | | Authors: | Miggiano, R, Casazza, V, Garavaglia, S, Ciaramella, M, Perugino, G, Rizzii, M, Rossi, F. | | Deposit date: | 2013-04-02 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and Structural Studies of the Mycobacterium Tuberculosis O6-Methylguanine Methyltransferase and Mutated Variants.
J.Bacteriol., 195, 2013
|
|
4BF8
 
 | | Fpr4 PPI domain | | Descriptor: | FPR4 | | Authors: | Monneau, Y, Mackereth, C. | | Deposit date: | 2013-03-15 | | Release date: | 2013-07-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and Activity of the Peptidyl-Prolyl Isomerase Domain from the Histone Chaperone Fpr4 Towards Histone H3 Proline Isomerization
J.Biol.Chem., 288, 2013
|
|
5EDK
 
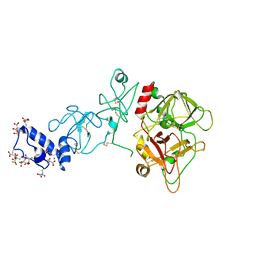 | | Crystal structure of prothrombin deletion mutant residues 146-167 ( Form II ). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | How the Linker Connecting the Two Kringles Influences Activation and Conformational Plasticity of Prothrombin.
J.Biol.Chem., 291, 2016
|
|
5W78
 
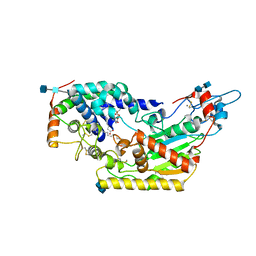 | | Human acyloxyacyl hydrolase (AOAH), proteolytically processed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acyloxyacyl hydrolase, CALCIUM ION, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | Crystal structure of the mammalian lipopolysaccharide detoxifier.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5WYN
 
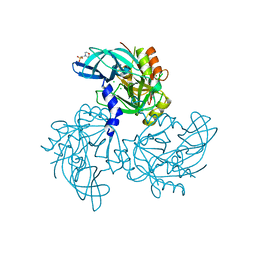 | | HtrA2 Pathogenic Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Serine protease HTRA2, ... | | Authors: | Wagh, A.R, Bose, K. | | Deposit date: | 2017-01-13 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of inactivation of human counterpart of mouse motor neuron degeneration 2 mutant in serine protease HtrA2
Biosci. Rep., 38, 2018
|
|
5W7C
 
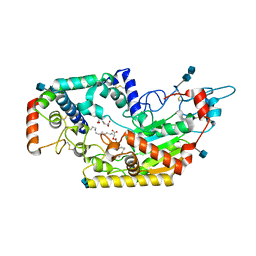 | | Human acyloxyacyl hydrolase (AOAH), proteolytically processed, S263A mutant, with LPS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-HYDROXY-TETRADECANOIC ACID, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of the mammalian lipopolysaccharide detoxifier.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1WO9
 
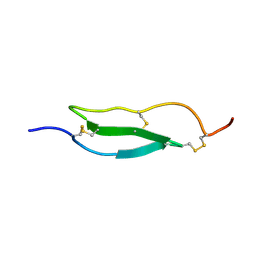 | | Selective inhibition of trypsins by insect peptides: role of P6-P10 loop | | Descriptor: | trypsin inhibitor | | Authors: | Kellenberger, C, Ferrat, G, Leone, P, Darbon, H, Roussel, A. | | Deposit date: | 2004-08-12 | | Release date: | 2004-09-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Selective inhibition of trypsins by insect peptides: role of P6-P10 loop
Biochemistry, 42, 2003
|
|
8FZZ
 
 | |
8G0K
 
 | |
4D7Z
 
 | | E. coli L-aspartate-alpha-decarboxylase mutant N72Q to a resolution of 1.9 Angstroms | | Descriptor: | ASPARTATE 1-DECARBOXYLASE ALPHA CHAIN, ASPARTATE 1-DECARBOXYLASE BETA CHAIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bravo, J.P.K, Monteiro, D.C.F, Webb, M.E, Pearson, A.R. | | Deposit date: | 2014-12-02 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the E. Coli L-Aspartate-Alpha-Decarboxylase Mutant N72Q to a Resolution of 1.9 Angstroms
To be Published
|
|
8FZY
 
 | |
6OC9
 
 | | S8 phosphorylated beta amyloid 40 fibrils | | Descriptor: | Amyloid-beta precursor protein, PHOSPHONATE | | Authors: | Qiang, W, Hu, Z.W. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Molecular structure of an N-terminal phosphorylated beta-amyloid fibril.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2L5Z
 
 | | NMR structure of the A730 loop of the Neurospora VS ribozyme | | Descriptor: | RNA (26-MER) | | Authors: | Desjardins, G, Bonneau, E, Girard, N, Boisbouvier, J, Legault, P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site.
Nucleic Acids Res., 39, 2011
|
|
3VH8
 
 | | KIR3DL1 in complex with HLA-B*5701 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B
Nature, 479, 2011
|
|
3TNX
 
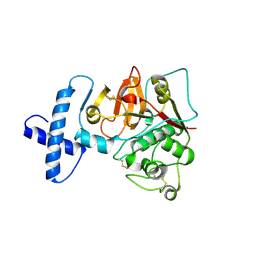 | | Structure of the precursor of a thermostable variant of papain at 2.6 Angstroem resolution | | Descriptor: | CHLORIDE ION, Papain | | Authors: | Roy, S, Choudhury, D, Dattagupta, J.K, Biswas, S. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structure of a thermostable mutant of pro-papain reveals its activation mechanism
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2JKH
 
 | | Factor Xa - cation inhibitor complex | | Descriptor: | 3-[(3~{a}~{S},4~{R},8~{a}~{S},8~{b}~{R})-4-[5-(5-chloranylthiophen-2-yl)-1,2-oxazol-3-yl]-1,3-bis(oxidanylidene)-4,6,7,8,8~{a},8~{b}-hexahydro-3~{a}~{H}-pyrrolo[3,4-a]pyrrolizin-2-yl]propyl-trimethyl-azanium, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Salonen, L.M, Bucher, C, Banner, D.W, Benz, J, Haap, W, Mary, J.L, Schweizer, W.B, Seiler, P, Kuster, O, Diederich, F. | | Deposit date: | 2008-08-28 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Cation-Pi Interactions at the Active Site of Factor Xa: Dramatic Enhancement Upon Stepwise N-Alkylation of Ammonium Ions.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2AXG
 
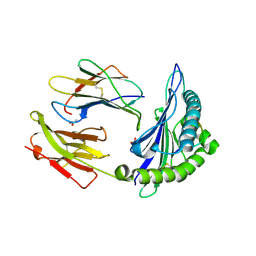 | | The Immunogenicity of a Viral Cytotoxic T Cell Epitope is controlled by its MHC-bound Conformation | | Descriptor: | 10-mer peptide from BZLF1 trans-activator protein, ACETIC ACID, Beta-2-microglobulin, ... | | Authors: | Tynan, F.E, Elhassen, D, Purcell, A.W, Burrows, J.M, Borg, N.A, Miles, J.J, Williamson, N.A, Green, K.J, Tellam, J, Kjer-Nielsen, L, McCluskey, J, Rossjohn, J, Burrows, S.R. | | Deposit date: | 2005-09-05 | | Release date: | 2005-11-29 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation
J.Exp.Med., 202, 2005
|
|
9EN6
 
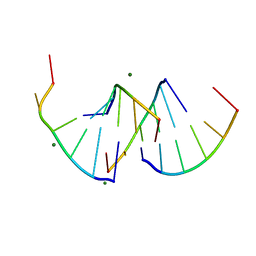 | | Crystal structure of RNA G2C4 repeats - native model pH 6.5 | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Mateja-Pluta, M, Kiliszek, A. | | Deposit date: | 2024-03-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (0.918 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
9CTH
 
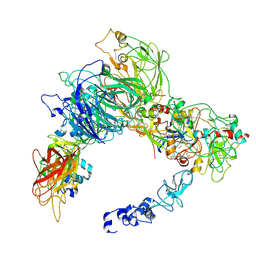 | | Preliminary map of the Prothrombin-prothrombinase complex on nano discs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activated Factor V (FVa) heavy chain, Activated Factor V (FVa) light chain, ... | | Authors: | Stojanovski, B.M, Mohammed, B.M, Di Cera, E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | The Prothrombin-Prothrombinase Interaction.
Subcell Biochem, 104, 2024
|
|
2AYV
 
 | | Crystal structure of a putative ubiquitin-conjugating enzyme E2 from Toxoplasma gondii | | Descriptor: | UNKNOWN ATOM OR ION, ubiquitin-conjugating enzyme E2 | | Authors: | Tempel, W, Dong, A, Zhao, Y, Lew, J, Alam, Z, Melone, M, Wasney, G, Kozieradzki, I, Vedadi, M, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-08 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
2AQU
 
 | | Structure of HIV-1 protease bound to atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, HIV-1 Protease | | Authors: | Clemente, J.C, Coman, R.M, Thiaville, M.M, Janka, L.K, Jeung, J.A, Nukoolkarn, S, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Leelamanit, W, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2005-08-18 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of HIV-1 CRF_01 A/E protease inhibitor resistance: structural determinants for maintaining sensitivity and developing resistance to atazanavir.
Biochemistry, 45, 2006
|
|
2AZR
 
 | | Crystal structure of PTP1B with Bicyclic Thiophene inhibitor | | Descriptor: | 3-(CARBOXYMETHOXY)THIENO[2,3-B]PYRIDINE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase, non-receptor type 1 | | Authors: | Moretto, A.F, Kirincich, S.J, Xu, W.X, Smith, M.J, Wan, Z.K, Wilson, D.P, Follows, B.C, Binnun, E, Joseph-McCarthy, D, Foreman, K, Erbe, D.V, Zhang, Y.L, Tam, S.K, Tam, S.Y, Lee, J. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bicyclic and tricyclic thiophenes as protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
