4YDF
 
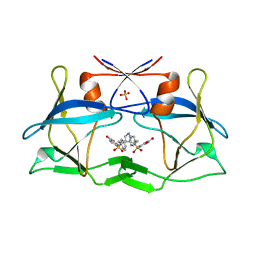 | | Crystal structure of compound 9 in complex with HTLV-1 Protease | | Descriptor: | HTLV-1 Protease, N-benzyl-N-[(3S,4S)-4-{benzyl[(4-nitrophenyl)sulfonyl]amino}pyrrolidin-3-yl]-3-nitrobenzenesulfonamide, SULFATE ION | | Authors: | Kuhnert, M, Blum, A, Steuber, H, Diederich, W.E. | | Deposit date: | 2015-02-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Privileged Structures Meet Human T-Cell Leukemia Virus-1 (HTLV-1): C2-Symmetric 3,4-Disubstituted Pyrrolidines as Nonpeptidic HTLV-1 Protease Inhibitors.
J.Med.Chem., 58, 2015
|
|
4YLC
 
 | |
2MOQ
 
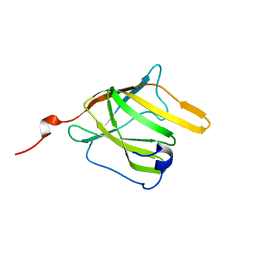 | | Solution Structure and Molecular determinants of Hemoglobin Binding of the first NEAT Domain of IsdB in Staphylococcus aureus | | Descriptor: | Iron-regulated surface determinant protein B | | Authors: | Fonner, B.A, Tripet, B.P, Eilers, B.J, Stanisich, J, Sullivan-Springhetti, R.K, Moore, R, Lui, M, Lei, B, Copie, V. | | Deposit date: | 2014-04-29 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Molecular Determinants of Hemoglobin Binding of the First NEAT Domain of IsdB in Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
5UIF
 
 | | Crystal Structure of Native Ps01740 | | Descriptor: | Ps01740 | | Authors: | LeVieux, J, Baas, B.J, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2017-01-13 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Kinetic and structural characterization of a cis-3-Chloroacrylic acid dehalogenase homologue in Pseudomonas sp. UW4: A potential step between subgroups in the tautomerase superfamily.
Arch. Biochem. Biophys., 636, 2017
|
|
4YMK
 
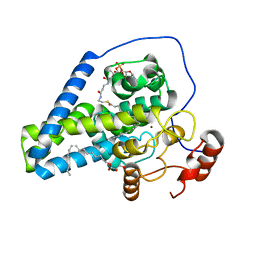 | | Crystal Structure of Stearoyl-Coenzyme A Desaturase 1 | | Descriptor: | Acyl-CoA desaturase 1, STEAROYL-COENZYME A, ZINC ION, ... | | Authors: | Bai, Y, McCoy, J.G, Rajashankar, K.R, Zhou, M. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | X-ray structure of a mammalian stearoyl-CoA desaturase.
Nature, 524, 2015
|
|
2MQP
 
 | |
2MU7
 
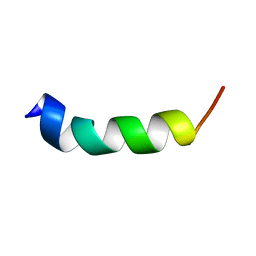 | | Shortening and modifying the 1513 MSP-1 peptide's alpha-helical region induces protection against malaria | | Descriptor: | 1513 MSP-1 peptide | | Authors: | Espejo, F, Bermudez, A, Torres, E, Urquiza, M, Rodriguez, R, Lopez, Y, Patarroyo, M. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Shortening and modifying the 1513 MSP-1 peptide's alpha-helical region induces protection against malaria.
Biochem.Biophys.Res.Commun., 315, 2004
|
|
5UKW
 
 | |
5UJ9
 
 | |
4YO1
 
 | |
5ULX
 
 | |
2MWG
 
 | |
4YIH
 
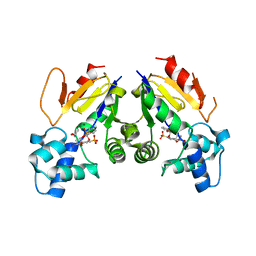 | | Crystal structure of human cytosolic 5'(3')-deoxyribonucleotidase in complex with the inhibitor PB-PVU | | Descriptor: | 1-{2-deoxy-3,5-O-[phenyl(phosphono)methylidene]-beta-D-threo-pentofuranosyl}-5-[(E)-2-phosphonoethenyl]pyrimidine-2,4(1H,3H)-dione, 5'(3')-deoxyribonucleotidase, cytosolic type, ... | | Authors: | Pachl, P, Rezacova, P, Brynda, J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-based design of a bisphosphonate 5'(3')-deoxyribonucleotidase inhibitor
Medchemcomm, 6, 2015
|
|
6BFZ
 
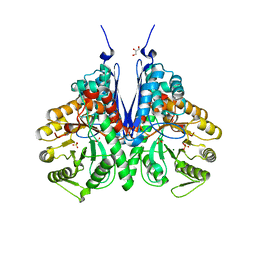 | | Crystal structure of enolase from E. coli with a mixture of apo form, substrate, and product form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PHOSPHOGLYCERIC ACID, Enolase, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
4YIR
 
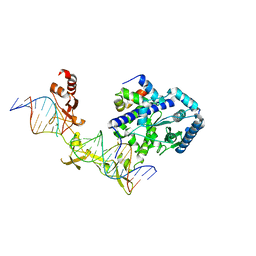 | | Crystal structure of Rad4-Rad23 crosslinked to an undamaged DNA | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*CP*G*GP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*CP*GP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Min, J.-H, Chen, X, Kim, Y. | | Deposit date: | 2015-03-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0501 Å) | | Cite: | Kinetic gating mechanism of DNA damage recognition by Rad4/XPC.
Nat Commun, 6, 2015
|
|
5UKP
 
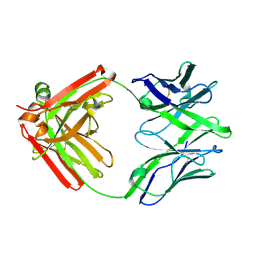 | | Structure of unliganded anti-gp120 CD4bs antibody DH522.1 Fab | | Descriptor: | DH522.1 Fab fragment heavy chain, DH522.1 Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
5UKN
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522UCA Fab | | Descriptor: | CHLORIDE ION, DH522UCA Fab fragment heavy chain, DH522UCA Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
4YQE
 
 | | Crystal structure of E. coli WrbA in complex with benzoquinone | | Descriptor: | 1,4-benzoquinone, FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone) | | Authors: | Degtjarik, O, Brynda, J, Ettrichova, O, Carey, J, Kuta Smatanova, I, Ettrich, R. | | Deposit date: | 2015-03-13 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Quantum Calculations Indicate Effective Electron Transfer between FMN and Benzoquinone in a New Crystal Structure of Escherichia coli WrbA.
J.Phys.Chem.B, 120, 2016
|
|
5V18
 
 | | Structure of PHD2 in complex with 1,2,4-Triazolo-[1,5-a]pyridine | | Descriptor: | 4-([1,2,4]triazolo[1,5-a]pyridin-5-yl)benzonitrile, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 1,2,4-Triazolo-[1,5-a]pyridine HIF Prolylhydroxylase Domain-1 (PHD-1) Inhibitors With a Novel Monodentate Binding Interaction.
J. Med. Chem., 60, 2017
|
|
5UOB
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with (R)-3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-(2-(methylamino)propyl)benzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(2R)-2-(methylamino)propyl]benzonitrile, CHLORIDE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
2NBZ
 
 | |
6B7A
 
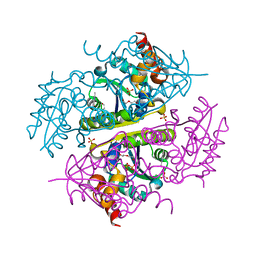 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-methyl-1H-benzo[d]imidazol-4-ol | | Descriptor: | 2-methyl-1H-benzimidazol-7-ol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
4Y3S
 
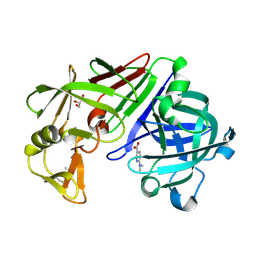 | |
4Y47
 
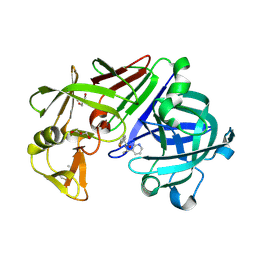 | | Endothiapepsin in complex with fragment 162 | | Descriptor: | 4-oxo-N-[(1S)-1-(pyridin-3-yl)ethyl]-4-(thiophen-2-yl)butanamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Krug, M, Uehlein, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
2N7A
 
 | | Solution structure of the human Siglec-8 lectin domain | | Descriptor: | Sialic acid-binding Ig-like lectin 8 | | Authors: | Proepster, J.M, Yang, F, Rabbani, S, Ernst, B, Allain, F.H.-T, Schubert, M. | | Deposit date: | 2015-09-07 | | Release date: | 2016-07-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
