7RMT
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-70 | | Descriptor: | 2-chloro-4-[4-(2,6-dioxo-1,2,5,6-tetrahydropyrimidine-4-carbonyl)piperazin-1-yl]benzaldehyde, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7AV8
 
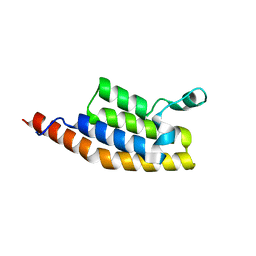 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212 | | Descriptor: | PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
7RNH
 
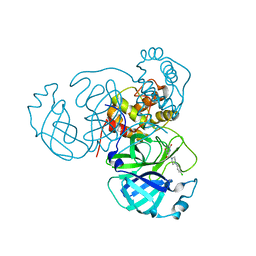 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-45 | | Descriptor: | 3C-like proteinase, 6-[4-(4-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
8A3A
 
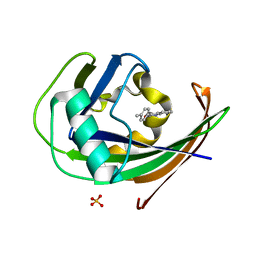 | | MTH1 in complex with TH013074 | | Descriptor: | 4-(4-methylphenyl)-1~{H}-quinazolin-2-one, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Liu, H, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-06-07 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 in complex with TH013074
To Be Published
|
|
7MGR
 
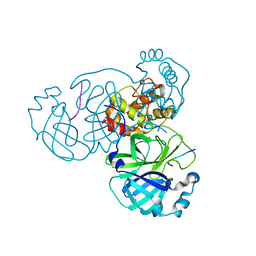 | |
7RLS
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-68 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4,5-trichlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7AWD
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG)in complex with garcinoic acid | | Descriptor: | (2Z,6E,10E)-13-[(2R)-6-hydroxy-2,8-dimethyl-3,4-dihydro-2H-1-benzopyran-2-yl]-2,6,10-trimethyltrideca-2,6,10-trienoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Endogenous vitamin E metabolites mediate allosteric PPAR gamma activation with unprecedented co-regulatory interactions.
Cell Chem Biol, 28, 2021
|
|
8A07
 
 | |
8A0T
 
 | | MTH1 in complex with TH012532 | | Descriptor: | 5-(phenylmethyl)pyrimidine-2,4-diol, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Scaletti, E, Stenmark, P. | | Deposit date: | 2022-05-30 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MTH1 in complex with TH012532
To Be Published
|
|
6D49
 
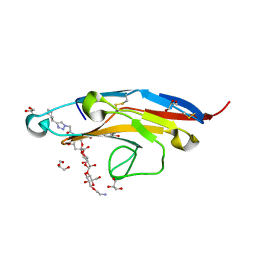 | | Cell Surface Receptor in Complex with Ligand at 1.80-A Resolution | | Descriptor: | 2-aminoethyl 5-{[(4-cyclohexyl-1H-1,2,3-triazol-1-yl)acetyl]amino}-3,5,9-trideoxy-9-[(4-hydroxy-3,5-dimethylbenzene-1-carbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl-(2->6)-beta-D-galactopyranosyl-(1->4)-beta-D-glucopyranoside, GLYCEROL, Myeloid cell surface antigen CD33 | | Authors: | Hermans, S.J, Miles, L.A, Parker, M.W. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Small Molecule Binding to Alzheimer Risk Factor CD33 Promotes A beta Phagocytosis.
Iscience, 19, 2019
|
|
6J0J
 
 | |
7ZJV
 
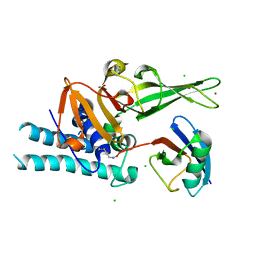 | |
7ZET
 
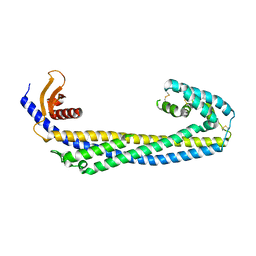 | | Crystal structure of human Clusterin, crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Clusterin, ... | | Authors: | Yuste-Checa, P, Bracher, A, Hartl, F.U. | | Deposit date: | 2022-03-31 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human Clusterin, crystal form I
To be published
|
|
7ZEU
 
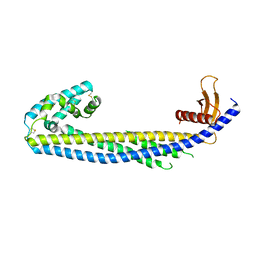 | |
7ZJU
 
 | |
6D50
 
 | | Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, CALCIUM ION, Glycosyl hydrolases family 2, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
8A34
 
 | | MTH1 in complex with TH013071 | | Descriptor: | 5-(2-phenylphenyl)-1H-pyrimidine-2,4-dione, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Liu, H, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-06-07 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MTH1 in complex with TH013071
To Be Published
|
|
8SYC
 
 | | Crystal structure of PDE3B in complex with GSK4394835A | | Descriptor: | MAGNESIUM ION, [3-[(4,7-dimethoxyquinolin-2-yl)carbonylamino]-5-[methyl-(phenylmethyl)carbamoyl]phenyl]-oxidanyl-oxidanylidene-boron, cGMP-inhibited 3',5'-cyclic phosphodiesterase 3B | | Authors: | Concha, N.O, Nolte, R. | | Deposit date: | 2023-05-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and SAR Study of Boronic Acid-Based Selective PDE3B Inhibitors from a Novel DNA-Encoded Library.
J.Med.Chem., 67, 2024
|
|
7REO
 
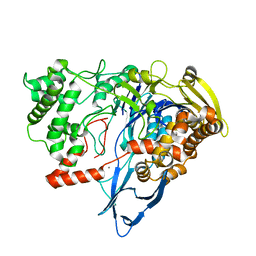 | |
7RMB
 
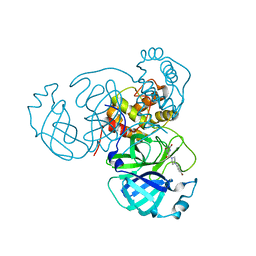 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-78 | | Descriptor: | 3C-like proteinase, 6-[4-(4-bromo-3-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7AVC
 
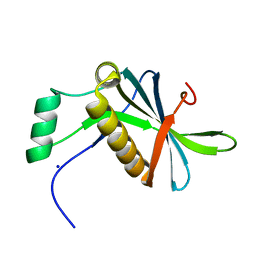 | | DoBi scaffold based on PIH1D1 N-terminal domain | | Descriptor: | GLYCEROL, PIH1 domain-containing protein 1, SODIUM ION | | Authors: | Kolenko, P, Pham, N.P, Pavlicek, J, Mikulecky, P, Schneider, B. | | Deposit date: | 2020-11-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein Binder (ProBi) as a New Class of Structurally Robust Non-Antibody Protein Scaffold for Directed Evolution.
Viruses, 13, 2021
|
|
7ZJ1
 
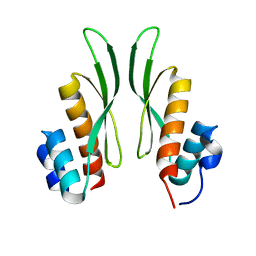 | | Crystal structure of ADAR1-dsRBD3 dimer | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Mboukou, A, Barraud, P. | | Deposit date: | 2022-04-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dimerization of ADAR1 modulates site-specificity of RNA editing
Biorxiv, 2023
|
|
6D4U
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 27 (VCC663664) | | Descriptor: | 2-(2,4-dimethoxyphenyl)-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
7RN4
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-69 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperidine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
6IZ4
 
 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | Trimeric intracellular cation channel type B-B | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
