4QOC
 
 | |
4V46
 
 | | Crystal structure of the BAFF-BAFF-R complex | | Descriptor: | MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Kim, H.M, Yu, K.S, Lee, M.E, Shin, D.R, Kim, Y.S, Paik, S.G, Yoo, O.J, Lee, H, Lee, J.-O. | | Deposit date: | 2003-03-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation
NAT.STRUCT.BIOL., 10, 2003
|
|
1PND
 
 | |
3WJX
 
 | | Wild-type orotidine 5'-monophosphate decarboxylase from M. thermoautotrophicus complexed with 6-amino-UMP | | Descriptor: | 6-AMINOURIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fujihashi, M, Kuroda, S, Pai, E.F, Miki, K. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Substrate distortion contributes to the catalysis of orotidine 5'-monophosphate decarboxylase.
J.Am.Chem.Soc., 135, 2013
|
|
3WJW
 
 | | Wild-type orotidine 5'-monophosphate decarboxylase from M. thermoautotrophicus complexed with 6-methyl-UMP | | Descriptor: | 6-methyluridine 5'-(dihydrogen phosphate), Orotidine 5'-phosphate decarboxylase | | Authors: | Fujihashi, M, Kuroda, S, Pai, E.F, Miki, K. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Substrate distortion contributes to the catalysis of orotidine 5'-monophosphate decarboxylase.
J.Am.Chem.Soc., 135, 2013
|
|
5A31
 
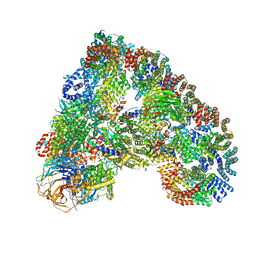 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
3K5K
 
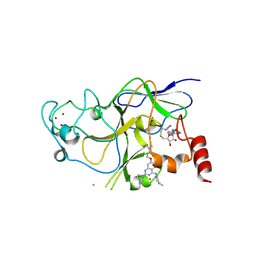 | | Discovery of a 2,4-Diamino-7-aminoalkoxy-quinazoline as a Potent Inhibitor of Histone Lysine Methyltransferase, G9a | | Descriptor: | 7-[3-(dimethylamino)propoxy]-6-methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinazolin-4-amine, CHLORIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Dong, A, Wasney, G.A, Liu, F, Chen, X, Allali-Hassani, A, Senisterra, G, Chau, I, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Frye, S.V, Bochkarev, A, Brown, P.J, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a.
J.Med.Chem., 52, 2009
|
|
3JVI
 
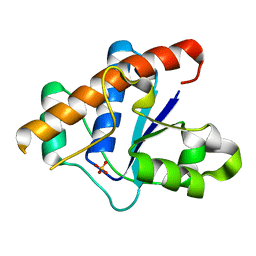 | |
2IJ7
 
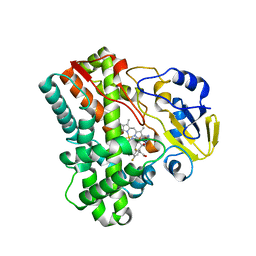 | | Structure of Mycobacterium tuberculosis CYP121 in complex with the antifungal drug fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roujeinikova, A, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis P450 CYP121-fluconazole complex reveals new azole drug-P450 binding mode.
J.Biol.Chem., 281, 2006
|
|
3WJY
 
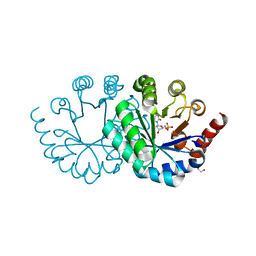 | |
6AQG
 
 | |
1V7R
 
 | |
2Y54
 
 | | Fragment growing induces conformational changes in acetylcholine- binding protein: A structural and thermodynamic analysis - (Fragment 1) | | Descriptor: | CHLORIDE ION, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION, ... | | Authors: | Rucktooa, P, Edink, E, deEsch, I.J.P, Sixma, T.K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
J.Am.Chem.Soc., 133, 2011
|
|
2Y56
 
 | | Fragment growing induces conformational changes in acetylcholine- binding protein: A structural and thermodynamic analysis - (Compound 3) | | Descriptor: | CHLORIDE ION, GLYCEROL, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Rucktooa, P, Edink, E, deEsch, I.J.P, Sixma, T.K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
J.Am.Chem.Soc., 133, 2011
|
|
5T71
 
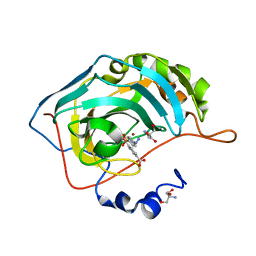 | | Human carboanhydrase F131C_C206S double mutant in complex with SA-2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(HYDROXYMERCURY)BENZOIC ACID, 4-[(E)-diazenyl]benzene-1-sulfonamide, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
2Y57
 
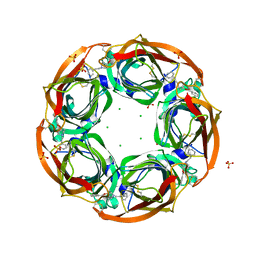 | | Fragment growing induces conformational changes in acetylcholine- binding protein: A structural and thermodynamic analysis - (Compound 4) | | Descriptor: | CHLORIDE ION, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION, ... | | Authors: | Rucktooa, P, Edink, E, deEsch, I.J.P, Sixma, T.K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
J.Am.Chem.Soc., 133, 2011
|
|
8E9N
 
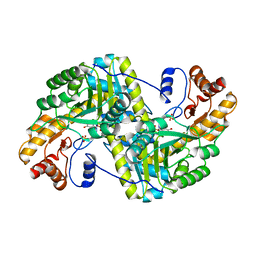 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
7N12
 
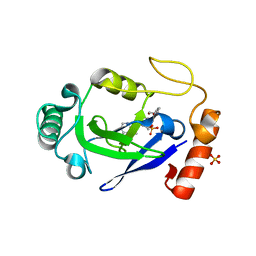 | | Crystal structure of the M. abscessus LeuRS editing domain in complex with epetraborole-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1S,5R,6R,7'S,8R)-7'-(aminomethyl)-6-(6-aminopurin-9-yl)-2'-(3-oxidanylpropoxy)spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,9'-8-oxa-9-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-8-yl]methyl dihydrogen phosphate | | Authors: | Kalthoff, E, Schmeing, M. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Efficacy of epetraborole against Mycobacterium abscessus is increased with norvaline.
Plos Pathog., 17, 2021
|
|
7N11
 
 | |
8E9L
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
4RDX
 
 | | Structure of histidinyl-tRNA synthetase in complex with tRNA(His) | | Descriptor: | ADENOSINE MONOPHOSPHATE, HISTIDINE, Histidine--tRNA ligase, ... | | Authors: | Xie, W, Tian, Q, Wang, C. | | Deposit date: | 2014-09-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for recognition of G-1-containing tRNA by histidyl-tRNA synthetase
Nucleic Acids Res., 43, 2015
|
|
2ONE
 
 | |
8E9J
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9P
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9C
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS in the ligand-free form at 100 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
