5WAQ
 
 | | Structure of BamD from Neisseria gonorrhoeae | | Descriptor: | Outer membrane protein assembly factor BamD | | Authors: | Korotkov, K.V, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and functional insights into the role of BamD and BamE within the beta-barrel assembly machinery in Neisseria gonorrhoeae.
J. Biol. Chem., 293, 2018
|
|
5WBK
 
 | |
5WKH
 
 | | D30 TCR in complex with HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, D30 TCR beta chain, GTS3 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WNS
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WQR
 
 | | High resolution structure of high-potential iron-sulfur protein in the reduced state | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Ohno, H, Takeda, K, Niwa, S, Tsujinaka, T, Hanazono, Y, Hirano, Y, Miki, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic characterization of the high-potential iron-sulfur protein in the oxidized state at 0.8 angstrom resolution
PLoS ONE, 12, 2017
|
|
5WKD
 
 | | Crystal structure of the segment, GNNQGSN, from the low complexity domain of TDP-43, residues 300-306 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Trinh, H, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WKF
 
 | | D30 TCR in complex with HLA-A*11:01-GTS1 | | Descriptor: | Beta-2-microglobulin, D30 TCR beta chain, GTS1 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WKL
 
 | | 1.85 A resolution structure of MERS 3CL protease in complex with piperidine-based peptidomimetic inhibitor 17 | | Descriptor: | (1R,2S)-2-{[N-({[4-benzyl-1-(tert-butoxycarbonyl)piperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[4-benzyl-1-(tert-butoxycarbonyl)piperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
5WLH
 
 | |
5X1E
 
 | |
5X1V
 
 | | PKM2 in complex with compound 2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-[2,3-bis(chloranyl)phenyl]carbonyl-1-methyl-pyrrole-2-carboxamide, Pyruvate kinase PKM | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2017-01-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and structure-guided fragment-linking of 4-(2,3-dichlorobenzoyl)-1-methyl-pyrrole-2-carboxamide as a pyruvate kinase M2 activator
Bioorg. Med. Chem., 25, 2017
|
|
5WLU
 
 | |
5WM4
 
 | | Crystal Structure of CahJ in Complex with 6-Methylsalicyl Adenylate | | Descriptor: | 9-(5-O-{(S)-hydroxy[(2-hydroxy-6-methylbenzene-1-carbonyl)oxy]phosphoryl}-alpha-L-lyxofuranosyl)-9H-purin-6-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5WMO
 
 | | Crystal Structure of HLA-B7 in complex with RPP, an EBV peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inability To Detect Cross-Reactive Memory T Cells Challenges the Frequency of Heterologous Immunity among Common Viruses.
J. Immunol., 200, 2018
|
|
5WN0
 
 | | APE1 exonuclease substrate complex with a C/G match | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular snapshots of APE1 proofreading mismatches and removing DNA damage.
Nat Commun, 9, 2018
|
|
5WN9
 
 | | Structure of antibody 2D10 bound to the central conserved region of RSV G | | Descriptor: | Major surface glycoprotein G, scFv 2D10 | | Authors: | Fedechkin, S.O, George, N.L, Wolff, J.T, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structures of respiratory syncytial virus G antigen bound to broadly neutralizing antibodies.
Sci Immunol, 3, 2018
|
|
5WNN
 
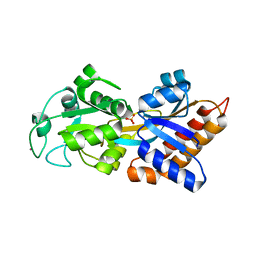 | |
3VGL
 
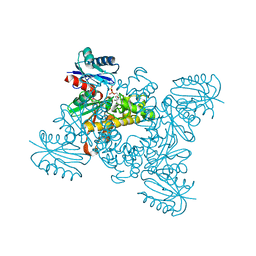 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus in complex with glucose and AMPPNP | | Descriptor: | Glucokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
5WB6
 
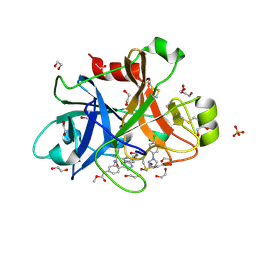 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5WBC
 
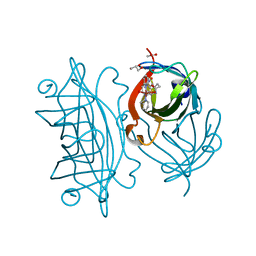 | | Designed Artificial Cupredoxins - WT | | Descriptor: | ACETATE ION, GLYCEROL, Streptavidin, ... | | Authors: | Mann, S.I, Heinisch, T, Weitz, A.C, Hendrich, M.R, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Modular Artificial Cupredoxins.
J. Am. Chem. Soc., 138, 2016
|
|
3VHM
 
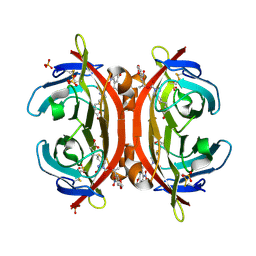 | | Crystal structure of NPC-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4R,6aR)-1-{[(1R)-1-(6-nitro-1,3-benzodioxol-5-yl)ethoxy]carbonyl}-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-29 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
5WO1
 
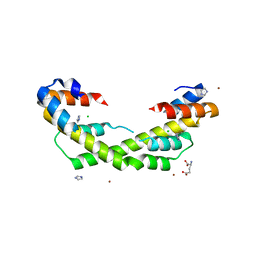 | | Chaperone Spy H96L bound to Im7 L18A L19A L37A (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, IMIDAZOLE, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WC8
 
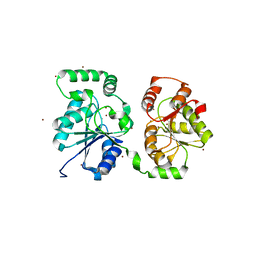 | |
5WP2
 
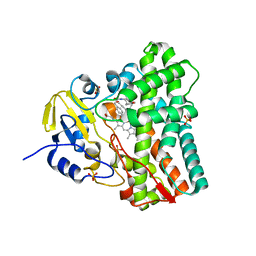 | | 1.44 Angstrom crystal structure of CYP121 from Mycobacterium tuberculosis in complex with substrate and CN | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, CYANIDE ION, Mycocyclosin synthase, ... | | Authors: | Fielding, A, Dornevil, K, Liu, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Probing Ligand Exchange in the P450 Enzyme CYP121 from Mycobacterium tuberculosis: Dynamic Equilibrium of the Distal Heme Ligand as a Function of pH and Temperature.
J. Am. Chem. Soc., 139, 2017
|
|
5WE8
 
 | |
