5F09
 
 | | Structure of inactive GCPII mutant in complex with beta-citryl glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Navratil, M, Pachl, P, Konvalinka, J. | | Deposit date: | 2015-11-27 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of human glutamate carboxypeptidases II and III reveals their divergent substrate specificities.
Febs J., 283, 2016
|
|
4EIG
 
 | | CA1698 camel antibody fragment in complex with DHFR | | Descriptor: | CA1698 camel antibody fragment, Dihydrofolate reductase | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
4EIZ
 
 | | Structure of Nb113 bound to apoDHFR | | Descriptor: | Dihydrofolate reductase, Nb113 Camel antibody fragment | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-06 | | Release date: | 2013-04-24 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
3KFY
 
 | | Dynamic switching and partial occupancies of a small molecule inhibitor complex of DHFR | | Descriptor: | 5-[(4-chlorophenyl)sulfanyl]quinazoline-2,4-diamine, CALCIUM ION, Dihydrofolate reductase, ... | | Authors: | Collins, E.J, Lee, A.L, Carroll, M.J, Gromova, A.V, Miller, K.R, Singleton, S.F. | | Deposit date: | 2009-10-28 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dynamic switching and partial occupancies of a small molecule inhibitor complex of DHFR
To be Published
|
|
8F7U
 
 | | Macrocyclic Plasmin Inhibitor | | Descriptor: | (5S,8R,18S,21R)-N-{[4-(aminomethyl)phenyl]methyl}-21-[(benzenesulfonyl)amino]-3,11,20-trioxo-2,5,8,12,19-pentaazatetracyclo[21.2.2.2~5,8~.2~13,16~]hentriaconta-1(25),13,15,23,26,28-hexaene-18-carboxamide, Activation peptide, GLYCEROL, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-11-20 | | Release date: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Synthesis and structural characterization of new macrocyclicplasmin inhibitors
Chemmedchem, 2023
|
|
1Z2J
 
 | |
3TSD
 
 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames complexed with XMP | | Descriptor: | D(-)-TARTARIC ACID, Inosine-5'-monophosphate dehydrogenase, SULFATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
8F7V
 
 | | Macrocyclic plasmin inhibitor | | Descriptor: | (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(benzenesulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, GLYCEROL, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-11-20 | | Release date: | 2023-02-08 | | Last modified: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthesis and structural characterization of new macrocyclicplasmin inhibitors
Chemmedchem, 2023
|
|
6WQX
 
 | | Human PRPK-TPRKB complex | | Descriptor: | EKC/KEOPS complex subunit TP53RK, EKC/KEOPS complex subunit TPRKB, MAGNESIUM ION, ... | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2020-04-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the human PRPK-TPRKB complex.
Commun Biol, 4, 2021
|
|
3TSB
 
 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | Descriptor: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
1Z5A
 
 | | Topoisomerase VI-B, ADP-bound dimer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
4EEV
 
 | | Crystal structure of c-Met in complex with LY2801653 | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[1-methyl-6-(1H-pyrazol-4-yl)-1H-indazol-5-yl]oxy}phenyl)-1-(4-fluorophenyl)-6-methyl-2-oxo-1,2-dihydropyridine-3-carboxamide | | Authors: | Wang, Y, Stout, S.L. | | Deposit date: | 2012-03-28 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models.
Invest New Drugs, 31, 2013
|
|
7Q83
 
 | |
2YGG
 
 | | Complex of CaMBR and CaM | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Koester, S, Yildiz, O. | | Deposit date: | 2011-04-15 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Structure of Human Na+/H+ Exchanger Nhe1 Regulatory Region in Complex with Cam and Ca2+
J.Biol.Chem., 286, 2011
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
8DJ4
 
 | | NMR Solution Structure of C-terminally amidated, Full-length Human Galanin | | Descriptor: | Galanin | | Authors: | Wilkinson, R.E, Kraichely, K.N, Buchanan, L.E, Parnham, S, Giuliano, M.W. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-07 | | Method: | SOLUTION NMR | | Cite: | The neuropeptide galanin adopts an irregular secondary structure.
Biochem.Biophys.Res.Commun., 626, 2022
|
|
5IJU
 
 | | Structure of an AA10 Lytic Polysaccharide Monooxygenase from Bacillus amyloliquefaciens with Cu(II) bound | | Descriptor: | 1,2-ETHANEDIOL, BaAA10 Lytic Polysaccharide Monooxygenase, CALCIUM ION, ... | | Authors: | Gregory, R.C, Hemsworth, G.R, Turkenburg, J.P, Hart, S.J, Walton, P.H, Davies, G.J. | | Deposit date: | 2016-03-02 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Activity, stability and 3-D structure of the Cu(ii) form of a chitin-active lytic polysaccharide monooxygenase from Bacillus amyloliquefaciens.
Dalton Trans, 45, 2016
|
|
5F0M
 
 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 and DMT1 (SeMet labeled) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Natural resistance-associated macrophage protein 2, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
5I8N
 
 | |
1HZK
 
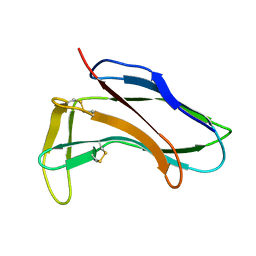 | |
2W6J
 
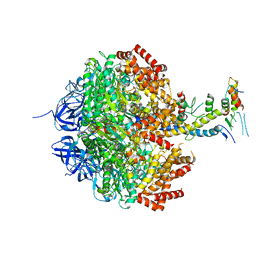 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 5. | | Descriptor: | ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT BETA, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Improving diffraction by humidity control: a novel device compatible with X-ray beamlines.
Acta Crystallogr. D Biol. Crystallogr., 65, 2009
|
|
3KPE
 
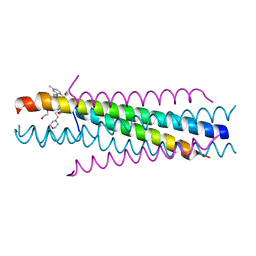 | | Solution structure of the respiratory syncytial virus (RSV)six-helix bundle complexed with TMC353121, a small-moleucule inhibitor of RSV | | Descriptor: | 2-[[6-[[[2-(3-hydroxypropyl)-5-methylphenyl]amino]methyl]-2-[[3-(4-morpholinyl)propyl]amino]-1H-benzimidazol-1-yl]methyl]-6-methyl-3-pyridinol, Fusion glycoprotein F0, TETRAETHYLENE GLYCOL | | Authors: | Roymans, D, De Bondt, H, Arnoult, E, Cummings, M.D, Van Vlijmen, H, Andries, K. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WEL
 
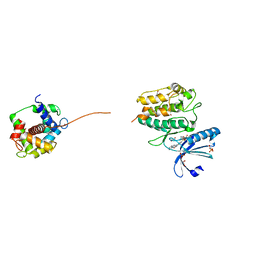 | | Crystal structure of SU6656-bound calcium/calmodulin-dependent protein kinase II delta in complex with calmodulin | | Descriptor: | (3Z)-N,N-DIMETHYL-2-OXO-3-(4,5,6,7-TETRAHYDRO-1H-INDOL-2-YLMETHYLIDENE)-2,3-DIHYDRO-1H-INDOLE-5-SULFONAMIDE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Rellos, P, Salah, E, Burgess-Brown, N, Keates, T, Muniz, J, Sethi, R, Roos, A, Filippakopoulos, P, von Delft, F, Edwards, A, Weigelt, J, Arrowsmith, C.H, Bountra, C, Knapp, S. | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Camkiidelta/Calmodulin Complex Reveals the Molecular Mechanism of Camkii Kinase Activation.
Plos Biol., 8, 2010
|
|
2WGW
 
 | | Crystal structure of the OXA-10 V117T mutant at pH 8.0 | | Descriptor: | BETA-LACTAMASE OXA-10, GLYCEROL, SULFATE ION | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
8I26
 
 | | NMR structure of Toxoplasma gondii PDCD5 (cis form) | | Descriptor: | Programmed cell death 5 protein | | Authors: | Lin, M.H, Hsu, C.H. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Proline Isomerization and Molten Globular Property of TgPDCD5 Secreted from Toxoplasma gondii Confers Its Regulation of Heparin Sulfate Binding.
Jacs Au, 4, 2024
|
|
