5F27
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 2 at 1.68A resolution | | Descriptor: | HTH-type transcriptional regulator EthR, ~{N}-methyl-1-(4-piperidin-1-ylphenyl)methanamine | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-12-01 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5F6H
 
 | |
5F04
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 3 at 1.84A resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-cyclopentyl-1-[3-[4-(methylaminomethyl)phenyl]-1,3-diazinan-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
3EUN
 
 | |
5F0O
 
 | | Cohesin subunit Pds5 in complex with Scc1 | | Descriptor: | KLTH0G16610p, cohesin subunit Pds5, KLTH0D07062p,KLTH0D07062p,KLTH0D07062p,cohesin subunit Pds5, ... | | Authors: | Lee, B.-G, Jansma, M, Nasmyth, K, Lowe, J. | | Deposit date: | 2015-11-27 | | Release date: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of the Cohesin Gatekeeper Pds5 and in Complex with Kleisin Scc1.
Cell Rep, 14, 2016
|
|
5F6J
 
 | |
5FDC
 
 | | Crystal structure of Human Carbonic Anhydrase II in complex with the anticonvulsant sulfamide JNJ-26990990 and its S,S-dioxide analog. | | Descriptor: | 3-[(sulfamoylamino)methyl]-1-benzothiophene, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Di Fiore, A, De Simone, G, Alterio, V, Riccio, V, Winum, J.-Y, Carta, F, Supuran, C.T. | | Deposit date: | 2015-12-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The anticonvulsant sulfamide JNJ-26990990 and its S,S-dioxide analog strongly inhibit carbonic anhydrases: solution and X-ray crystallographic studies.
Org.Biomol.Chem., 14, 2016
|
|
8VJU
 
 | |
8VJX
 
 | |
8VJV
 
 | |
8VH8
 
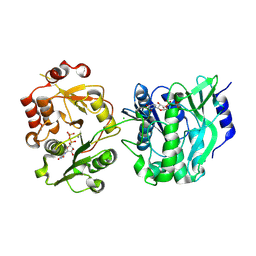 | | Crystal structure of heparosan synthase 2 from Pasteurella multocida at 2.85 A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pedersen, L.C, Liu, J, Stancanelli, E, krahn, J.M. | | Deposit date: | 2023-12-31 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Functional Analysis of Heparosan Synthase 2 from Pasteurella multocida to Improve the Synthesis of Heparin
Acs Catalysis, 14, 2024
|
|
5FHD
 
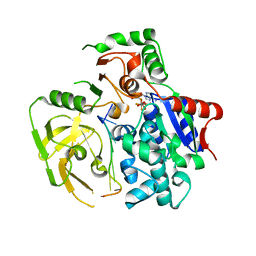 | | Structure of Bacteroides sp Pif1 complexed with tailed dsDNA resulting in ssDNA bound complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*CP*CP*GP*GP*GP*GP*CP*CP*GP*CP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FRP
 
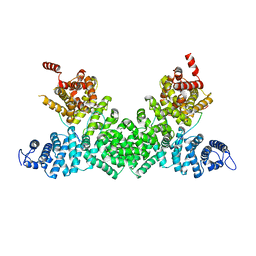 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | MCD1-LIKE PROTEIN, SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
5IUU
 
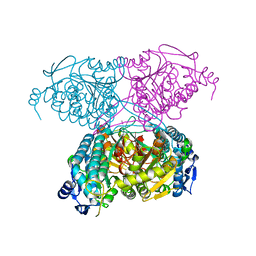 | |
5J61
 
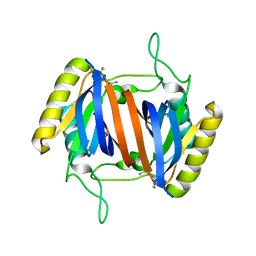 | |
8WA8
 
 | | Human transketolase in complex with phosphite | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PHOSPHITE ION, ... | | Authors: | Liu, Z, Tittmann, K, Dai, S. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
5FHF
 
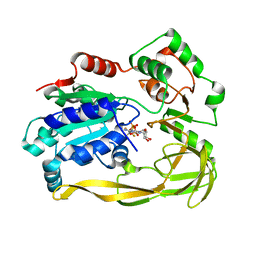 | | Crystal structure of Bacteroides sp Pif1 in complex with ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TETRAFLUOROALUMINATE ION, Uncharacterized protein | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
8W6X
 
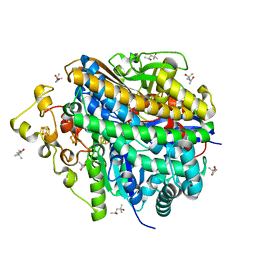 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
5FSG
 
 | |
8W85
 
 | | HLA-DQ2.5-gamma2 gliadin peptide in complex with DQN0385AE01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE01 Fab heavy chain, DQN0385AE01 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W86
 
 | | HLA-DQ2.5-B/C hordein peptide in complex with DQN0385AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0385AE02 Fab heavy chain, DQN0385AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
8W84
 
 | | HLA-DQ2.5-alpha2 gliadin peptide in complex with DQN0344AE02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQN0344AE02 Fab heavy chain, DQN0344AE02 Fab light chain, ... | | Authors: | Irie, M, Tsushima, T, Teranishi-Ikawa, Y, Takahashi, N, Ishii, S, Okura, Y, Fukami, T.A, Torizawa, T. | | Deposit date: | 2023-08-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease.
Nat Commun, 14, 2023
|
|
5FQ1
 
 | |
5IUV
 
 | | Crystal Structure of Indole-3-acetaldehyde Dehydrogenase in complexed with NAD+ | | Descriptor: | Aldehyde dehydrogenase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, S.G, McClerklin, S, Kunkel, B, Jez, J.M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000.
PLoS Pathog., 14, 2018
|
|
3E6Z
 
 | |
