1V0E
 
 | | Endosialidase of Bacteriophage K1F | | Descriptor: | ENDO-ALPHA-SIALIDASE, PHOSPHATE ION | | Authors: | Stummeyer, K, Dickmanns, A, Muehlenhoff, M, Gerady-Schahn, R, Ficner, R. | | Deposit date: | 2004-03-28 | | Release date: | 2004-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Polysialic Acid-Degrading Endosialidase of Bacteriophage K1F
Nat.Struct.Mol.Biol., 12, 2005
|
|
5K9R
 
 | | PDE10a with imidazopyrazine inhibitor | | Descriptor: | 4-[5-[1-(2-methoxyethyl)pyrazol-4-yl]-2-(quinolin-2-yloxymethyl)imidazo[1,2-a]pyrazin-8-yl]morpholine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gibbs, A.G, Schubert, C. | | Deposit date: | 2016-06-01 | | Release date: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of novel potent imidazo[1,2-b]pyridazine PDE10a inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5KAJ
 
 | | Crystal structure of a dioxygenase in the Crotonase superfamily in P21, A319C mutant | | Descriptor: | (3,5-dihydroxyphenyl)acetyl-CoA 1,2-dioxygenase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[(~{E})-2-[3,5-bis(oxidanyl)phenyl]-1-oxidanyl-ethenyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Li, K, Fielding, E.N, Condurso, H.L, Bruner, S.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Probing the structural basis of oxygen binding in a cofactor-independent dioxygenase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1KR1
 
 | | Hevamine Mutant D125A/E127A in Complex with Tetra-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevamine A, SULFATE ION | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression and Characterization of Active Site Mutants of Hevamine, a
Chitinase from the Rubber Tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
5KBY
 
 | | Crystal structure of dipeptidyl peptidase IV in complex with SYR-472 | | Descriptor: | 2-[[6-[(3~{R})-3-azanylpiperidin-1-yl]-3-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]methyl]-4-fluoranyl-benzenecarboni trile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skene, R.J, Jennings, A.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Trelagliptin (SYR-472, Zafatek), Novel Once-Weekly Treatment for Type 2 Diabetes, Inhibits Dipeptidyl Peptidase-4 (DPP-4) via a Non-Covalent Mechanism.
Plos One, 11, 2016
|
|
5JRQ
 
 | | BRAFV600E Kinase Domain In Complex with Chemically Linked Vemurafenib Inhibitor VEM-6-VEM | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{2,4-difluoro-3-[5-(4-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]phenyl}propane-1-sulfonamide, ... | | Authors: | Grasso, M.J, Marmorstein, R. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Chemically Linked Vemurafenib Inhibitors Promote an Inactive BRAF(V600E) Conformation.
Acs Chem.Biol., 11, 2016
|
|
5L1H
 
 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with noncompetitive inhibitor GYKI53655 | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
1UOF
 
 | | Deacetoxycephalosporin C synthase complexed with Penicillin G | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, PENICILLIN G | | Authors: | Valegard, K, Terwisscha Van scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1VAG
 
 | | Neuronal nitric oxide synthase oxygenase domain complexed with the inhibitor AR-R17477 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-(4-{2-[(3-CHLOROBENZYL)AMINO]ETHYL}PHENYL)THIOPHENE-2-CARBOXIMIDAMIDE, Nitric-oxide synthase, ... | | Authors: | Fedorov, R, Vasan, R, Ghosh, D.K, Schlichting, I. | | Deposit date: | 2004-02-16 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of nitric oxide synthase isoforms complexed with the inhibitor AR-R17477 suggest a rational basis for specificity and inhibitor design
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1UW9
 
 | | L290F-A222T chlamydomonas Rubisco mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2004-02-03 | | Release date: | 2005-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Altered Intersubunit Interactions in Crystal Structures of Catalytically Compromised Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase
Biochemistry, 44, 2005
|
|
1UVC
 
 | | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza sativa | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, STEARIC ACID | | Authors: | Cheng, H.-C, Cheng, P.-T, Peng, P, Lyu, P.-C, Sun, Y.-J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipid Binding in Rice Nonspecific Lipid Transfer Protein-1 Complexes from Oryza Sativa
Protein Sci., 13, 2004
|
|
1KI8
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-BROMOVINYLDEOXYURIDINE | | Descriptor: | 5-BROMOVINYLDEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
1UPP
 
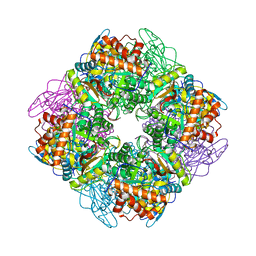 | | SPINACH RUBISCO IN COMPLEX WITH 2-CARBOXYARABINITOL 2 BISPHOSPHATE and Calcium. | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CALCIUM ION, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN, ... | | Authors: | Karkehabadi, S, Taylor, T.C, Andersson, I. | | Deposit date: | 2003-10-09 | | Release date: | 2003-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium Supports Loop Closure But not Catalysis in Rubisco
J.Mol.Biol., 334, 2003
|
|
1UP7
 
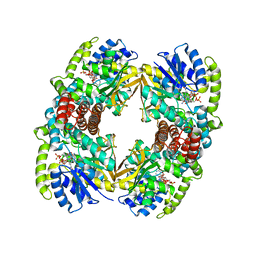 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.4 Angstrom resolution in the tetragonal form with NAD and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nad+ and Metal-Ion Dependent Hydrolysis by Family 4 Glycosidases: Structural Insight Into Specificity for Phospho-Beta-D-Glucosides
J.Mol.Biol., 346, 2005
|
|
1UR8
 
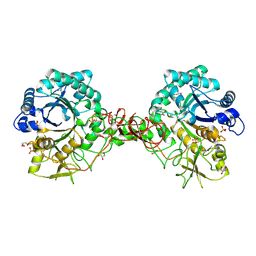 | | Interactions of a family 18 chitinase with the designed inhibitor HM508, and its degradation product, chitobiono-delta-lactone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, CHITINASE B, GLYCEROL, ... | | Authors: | Vaaje-Kolstad, G, Vasella, A, Peter, M.G, Netter, C, Houston, D.R, Westereng, B, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2003-10-27 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of a Family 18 Chitinase with the Designed Inhibitor Hm508 and its Degradation Product, Chitobiono-Delta-Lactone.
J.Biol.Chem., 279, 2004
|
|
1UTD
 
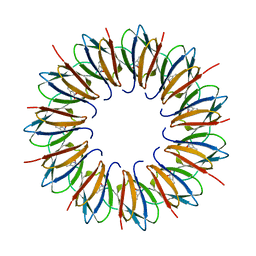 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a 63-nucleotide RNA molecule containing GAGUUU repeats | | Descriptor: | 5'-R(*GP*UP*UP*UP*GP*AP)-3', TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Hopcroft, N.H, Manfredo, A, Wendt, A.L, Brzozowski, A.M, Gollnick, P, Antson, A.A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Interaction of RNA with Trap: The Role of Triplet Repeats and Separating Spacer Nucleotides
J.Mol.Biol., 338, 2004
|
|
5KN5
 
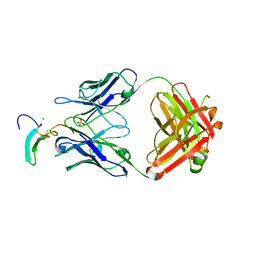 | | TGFalpha/Epiregulin complex with neutralizing antibody LY3016859 | | Descriptor: | Epiregulin Antibody LY3016859 Fab Heavy Chain, Epiregulin Antibody LY3016859 Fab Light Chain, Protransforming growth factor alpha, ... | | Authors: | Atwell, S, Boyles, J.S, Clawson, D.K, Druzina, Z, Josef, G.H, Weichert, K, Witcher, D.R. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-31 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of selectivity and neutralizing activity of a TGF alpha /epiregulin specific antibody.
Protein Sci., 25, 2016
|
|
1KJM
 
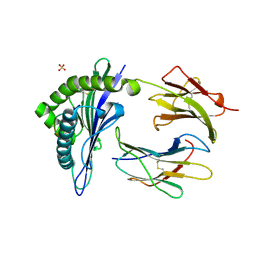 | | TAP-A-associated rat MHC class I molecule | | Descriptor: | B6 Peptide, RT1 class I histocompatibility antigen, AA alpha chain, ... | | Authors: | Rudolph, M.G, Stevens, J, Speir, J.A, Trowsdale, J, Butcher, G.W, Joly, E, Wilson, I.A. | | Deposit date: | 2001-12-04 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially
with peptide transporter alleles TAP-A and TAP-B.
J.Mol.Biol., 324, 2002
|
|
1UZ8
 
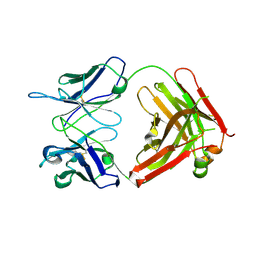 | | anti-Lewis X Fab fragment in complex with Lewis X | | Descriptor: | IGG FAB (IGG3, KAPPA) HEAVY CHAIN 291-2G3-A, KAPPA) LIGHT CHAIN 291-2G3-A, ... | | Authors: | Van Roon, A.M.M, Pannu, N.S, De Vrind, J.P.M, Hokke, C.H, Deelder, A.M, Van Der marel, G.A, Van Boom, J.H, Abrahams, J.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an Anti-Lewis X Fab Fragment in Complex with its Lewis X Antigen
Structure, 12, 2004
|
|
1V38
 
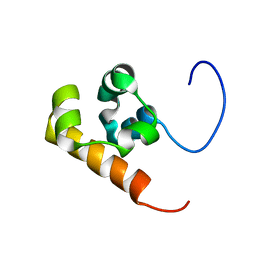 | | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1 | | Descriptor: | SAM-domain protein SAMSN-1 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-29 | | Release date: | 2004-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1
To be Published
|
|
5L5G
 
 | | Plexin A2 full extracellular region, domains 1 to 8 modeled, data to 10 angstrom | | Descriptor: | Plexin-A2 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5KR7
 
 | | KDM4C bound to pyrazolo-pyrimidine scaffold | | Descriptor: | 6-ethyl-2,5-dimethyl-7-oxidanylidene-4~{H}-pyrazolo[1,5-a]pyrimidine-3-carbonitrile, CHLORIDE ION, FE (II) ION, ... | | Authors: | Bellon, S.F, Poy, F, Setser, J.W. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of potent, selective KDM5 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1V6F
 
 | | Solution Structure of Glia Maturation Factor-beta from Mus Musculus | | Descriptor: | glia maturation factor, beta | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-29 | | Release date: | 2004-05-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
5KSQ
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5L7P
 
 | | In silico-powered specific incorporation of photocaged Dopa at multiple protein sites | | Descriptor: | (2~{S})-2-azanyl-3-[3-[(2-nitrophenyl)methoxy]-4-oxidanyl-phenyl]propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hauf, M, Richter, F, Schneider, T, Martins, B.M, Baumann, T, Durkin, P, Dobbek, H, Moeglich, A, Budisa, N. | | Deposit date: | 2016-06-03 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Photoactivatable Mussel-Based Underwater Adhesive Proteins by an Expanded Genetic Code.
Chembiochem, 18, 2017
|
|
