3KOS
 
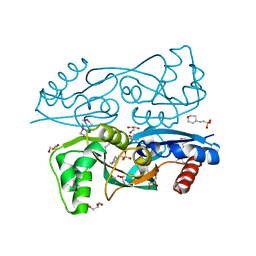 | |
5H2A
 
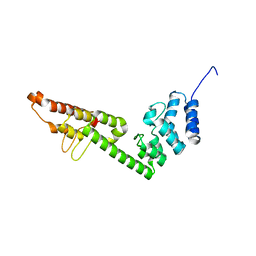 | |
6OS0
 
 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to angiotensin II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensinogen, CHLORIDE ION, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
5H2P
 
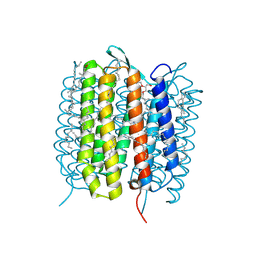 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 657 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
3KOZ
 
 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Anaerobic) | | Descriptor: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KUT
 
 | |
5H32
 
 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 5.0 | | Descriptor: | C10 IgG heavy chain variable region, C10 IgG light chain variable region, structural protein E | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lok, S.-M. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
3KQ6
 
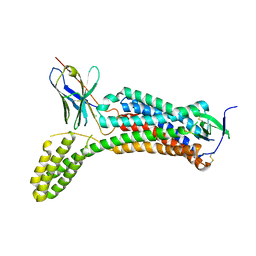
 | | Enhancing the Therapeutic Properties of a Protein by a Designed Zinc-Binding Site, Structural principles of a novel long-acting insulin analog | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Wan, Z.L, Hu, S.Q, Whittaker, L, Phillips, N.B, Whittake, J, Ismail-Beigi, F, Weiss, M.A. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Supramolecular protein engineering: design of zinc-stapled insulin hexamers as a long acting depot.
J.Biol.Chem., 285, 2010
|
|
5GSV
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-5 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GT4
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with (1R,2S,3R,5Z,7E,14beta,17alpha)-2-cyanopropoxy-9,10-secocholesta-5,7,10-triene-1,3,25-triol | | Descriptor: | 4-{[(1R,2S,3R,5Z,7E,14beta,17alpha)-1,3,25-trihydroxy-9,10-secocholesta-5,7,10-trien-2-yl]oxy}butanenitrile, Vitamin D3 receptor | | Authors: | Takimoto-Kamimura, M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the human vitamin D receptor ligand binding domain complexed with (1R,2S,3R,5Z,7E,14beta,17alpha)-2-cyanopropoxy-9,10-secocholesta-5,7,10-triene-1,3,25-triol
To Be Published
|
|
5H3G
 
 | | Crystal Structure of Oryza sativa Acyl-CoA-Binding Protein 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative Acyl-CoA binding protein (ACBP) | | Authors: | Kong, G.K.W, Chan, W.H.Y. | | Deposit date: | 2016-10-24 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The first plant acyl-CoA-binding protein structures: the close homologues OsACBP1 and OsACBP2 from rice
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3KV0
 
 | | Crystal structure of HET-C2: A FUNGAL GLYCOLIPID TRANSFER PROTEIN (GLTP) | | Descriptor: | HET-C2 | | Authors: | Simanshu, D.K, Kenoth, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2009-11-28 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determination and tryptophan fluorescence of heterokaryon incompatibility C2 protein (HET-C2), a fungal glycolipid transfer protein (GLTP), provide novel insights into glycolipid specificity and membrane interaction by the GLTP fold.
J.Biol.Chem., 285, 2010
|
|
5GT9
 
 | | The X-ray structure of 7beta-hydroxysteroid dehydrogenase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Wang, F, Wang, R, Lv, Z, Chen, Q, Huo, X. | | Deposit date: | 2016-08-19 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of NADP(+)-bound 7 beta-hydroxysteroid dehydrogenase reveals two cofactor-binding modes
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3KVA
 
 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5H4D
 
 | | Crystal structure of hSIRT3 in complex with a specific agonist Amiodarone hydrochloride | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 7-AMINO-4-METHYL-CHROMEN-2-ONE, ARG-HIS-LYS, ... | | Authors: | Zhang, S, Fu, L, Liu, J, Liu, B. | | Deposit date: | 2016-10-31 | | Release date: | 2017-11-08 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of hSIRT3 in complex with a specific agonist Amiodarone hydrochloride
To Be Published
|
|
5GTG
 
 | | Crystal structure of onion lachrymatory factor synthase (LFS) containing 1,2-propanediol | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lachrymatory-factor synthase, S-1,2-PROPANEDIOL, ... | | Authors: | Arakawa, T, Sato, Y, Takabe, J, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2016-08-20 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase
Acs Catalysis, 10, 2020
|
|
3KRG
 
 | |
5H54
 
 | | Mdm12 from K. lactis 1-239 | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S, Endo, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H59
 
 | | Ferredoxin-NADP+ reductase from maize root | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Kurisu, G, Hase, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP(+) oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation
Photosyn. Res., 134, 2017
|
|
5GTY
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with LXX-6-26 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(6-methylpyridin-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery and characterization of a novel irreversible EGFR mutants selective and potent kinase inhibitor CHMFL-EGFR-26 with a distinct binding mode.
Oncotarget, 8, 2017
|
|
3KRR
 
 | | Crystal Structure of JAK2 complexed with a potent quinoxaline ATP site inhibitor | | Descriptor: | 8-[3,5-difluoro-4-(morpholin-4-ylmethyl)phenyl]-2-(1-piperidin-4-yl-1H-pyrazol-4-yl)quinoxaline, Tyrosine-protein kinase JAK2 | | Authors: | Tavares, G.A, Gerspacher, M, Kroemer, M, Scheufler, C. | | Deposit date: | 2009-11-19 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and Selective Inhibition of Polycythemia by the Quinoxaline JAK2 Inhibitor NVP-BSK805
Mol.Cancer Ther., 9, 2010
|
|
5H5E
 
 | |
5GIZ
 
 | | Periplasmic heme-binding protein BhuT in apo form | | Descriptor: | CHLORIDE ION, Putative hemin transport system, substrate-binding protein, ... | | Authors: | Nakamura, N, Naoe, Y, Rahman, M.M, Shiro, Y, Sugimoto, H. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems.
Proteins, 85, 2017
|
|
3KRX
 
 | |
5GUX
 
 | | Cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with xenon | | Descriptor: | Antibody fab fragment heavy chain, Antibody fab fragment light chain, CALCIUM ION, ... | | Authors: | Ishii, S, Terasaka, E, Sugimoto, H, Shiro, Y, Tosha, T. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamics of nitric oxide controlled by protein complex in bacterial system.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
