5H2J
 
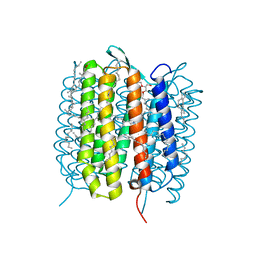 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 290 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
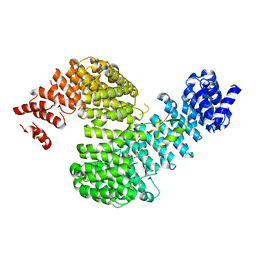
5H2V
 
 | |
5GRG
 
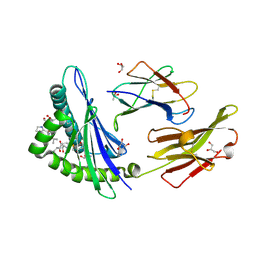 | | Crystal structure of dual peptide from EBV in complex with HLA-A*11:01 | | Descriptor: | Beta-2-microglobulin, Epstein Barr Virus, Latent Membrane Protein 2 epitope, ... | | Authors: | Tadwal, V.S, Xiao, Z, Ren, E.C. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dual non-contiguous peptide occupancy of HLA class I evoke antiviral human CD8 T cell response and form neo-epitopes with self-antigens
Sci Rep, 7, 2017
|
|
3KSB
 
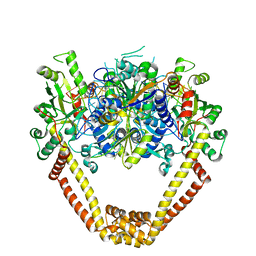 | | Detailed structural insight into the DNA cleavage complex of type IIA topoisomerases (re-sealed form) | | Descriptor: | 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*TP*GP*AP*CP*TP*AP*TP*GP*CP*AP*CP*GP*TP*AP*AP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*TP*AP*GP*TP*CP*AP*TP*TP*CP*AP*TP*GP*AP*CP*CP*TP*TP*GP*GP*T)-3', DNA topoisomerase 4 subunit A, ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2009-11-21 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
5H4G
 
 | | Structure of PIN-domain protein (VapC4 toxin) from Pyrococcus horikoshii determined at 1.77 A resolution | | Descriptor: | Ribonuclease VapC4, ZINC ION | | Authors: | Biswas, A, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure
J. Struct. Biol., 197, 2017
|
|
5H4S
 
 | |
5GRO
 
 | | Crystal structure of the N-terminal anticodon-binding domain of non-discriminating aspartyl-tRNA synthetase from Helicobacter pylori | | Descriptor: | Aspartate--tRNA(Asp/Asn) ligase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Songsiriritthigul, C, Fuengfuloy, P, Chen, C.-J, Suebka, S, Chuawong, P. | | Deposit date: | 2016-08-12 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the N-terminal anticodon-binding domain of the nondiscriminating aspartyl-tRNA synthetase from Helicobacter pylori
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5H57
 
 | | Ferredoxin III from maize root | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-3, chloroplastic | | Authors: | Kurisu, G, Hase, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP(+) oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation
Photosyn. Res., 134, 2017
|
|
5H5G
 
 | | Staphylococcus aureus FtsZ-GDP in T and R states | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
5GRW
 
 | | Crystal structure of homo-specific diabody | | Descriptor: | homo-specific diabody protein | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
3KUV
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - T42S mutant in complex with acetate. | | Descriptor: | ACETATE ION, Fluoroacetyl coenzyme A thioesterase | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
5GSA
 
 | | EED in complex with an allosteric PRC2 inhibitor | | Descriptor: | Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of
Nat. Chem. Biol., 13, 2017
|
|
5GG4
 
 | | Crystal structure of USP7 with RNF169 peptide | | Descriptor: | Peptide from E3 ubiquitin-protein ligase RNF169, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Jiang, Y, Gong, Q. | | Deposit date: | 2016-06-15 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Dual-utility NLS drives RNF169-dependent DNA damage responses.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3L35
 
 | | PIE12 D-peptide against HIV entry | | Descriptor: | GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
3KV4
 
 | | Structure of PHF8 in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, Histone H3-like, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5GGA
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-GDP, 8-oxo-GMP and pyrophosphate | | Descriptor: | Hydrolase, NUDIX family protein, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3L3I
 
 | | Crystal structure of HLA-B*4402 in complex with the F7A mutant of a self-peptide derived from DPA*0201 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5GSM
 
 | | Glycoside hydrolase B with product | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Watanabe, M, Kamachi, S, Mine, S. | | Deposit date: | 2016-08-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Glycoside hydrolase B with product
To Be Published
|
|
5GGD
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with phosphate and magnesium ions (excess magnesium, II) | | Descriptor: | Hydrolase, NUDIX family protein, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3KVS
 
 | |
5GTJ
 
 | |
5GGK
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Man-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-mannopyranoside, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3KWA
 
 | | Polyamines inhibit carbonic anhydrases | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, SPERMINE, ... | | Authors: | Temperini, C. | | Deposit date: | 2009-12-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Polyamines inhibit carbonic anhydrases by anchoring to the zinc-coordinated water molecule
J.Med.Chem., 53, 2010
|
|
5GTT
 
 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a | | Descriptor: | 1,2-ETHANEDIOL, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
5GGO
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GalNac-beta1,3-GlcNAc-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
