5IEV
 
 | | Crystal structure of BAY 1000394 (Roniciclib) bound to CDK2 | | Descriptor: | Cyclin-dependent kinase 2, Roniciclib | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
5IB4
 
 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR and Nickel | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
1IQP
 
 | | Crystal Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RFCS | | Authors: | Oyama, T, Ishino, Y, Cann, I.K.O, Ishino, S, Morikawa, K. | | Deposit date: | 2001-07-24 | | Release date: | 2001-09-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus
Mol.Cell, 8, 2001
|
|
5I17
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV1-69/IGKV3-20 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
1IT4
 
 | | Solution structure of the prokaryotic Phospholipase A2 from Streptomyces violaceoruber | | Descriptor: | CALCIUM ION, phospholipase A2 | | Authors: | Ohtani, K, Sugiyama, M, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-08 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.BIOL.CHEM., 277, 2002
|
|
1TG2
 
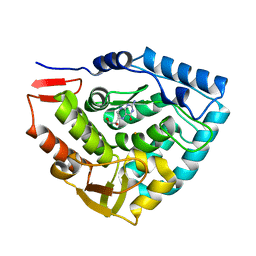 | | Crystal structure of phenylalanine hydroxylase A313T mutant with 7,8-dihydrobiopterin bound | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5I9M
 
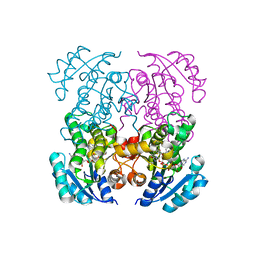 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT408 | | Descriptor: | 5-ethyl-4-fluoro-2-[(2-methylpyridin-3-yl)oxy]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5IB2
 
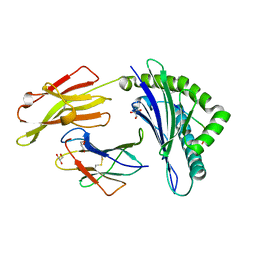 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
5IBE
 
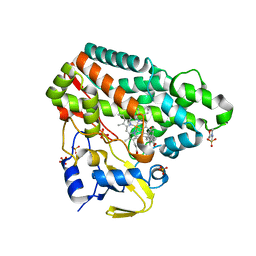 | |
1T5M
 
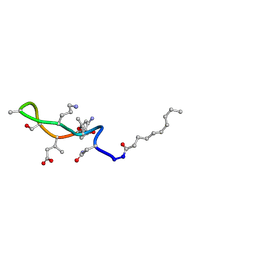 | | Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin | | Descriptor: | DAPTOMYCIN, DECANOIC ACID | | Authors: | Jung, D, Rozek, A, Okon, M, Hancock, R.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Structural Transitions as Determinants of the Action of the Calcium-Dependent Antibiotic Daptomycin.
Chem.Biol., 11, 2004
|
|
5IAT
 
 | |
5IB5
 
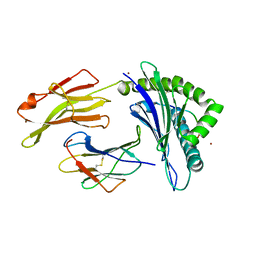 | | Crystal structure of HLA-B*27:09 complexed with the self-peptide pVIPR and Copper | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
5IBI
 
 | |
1IPP
 
 | | HOMING ENDONUCLEASE/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON-ENCODED ENDONUCLEASE I-PPOI, ... | | Authors: | Flick, K.E, Jurica, M.S, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 1998-03-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI.
Nature, 394, 1998
|
|
1IT5
 
 | | Solution structure of apo-type PLA2 from Streptomyces violaceruber A-2688. | | Descriptor: | Phospholipase A2 | | Authors: | Sugiyama, M, Ohtani, K, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-09 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.Biol.Chem., 277, 2002
|
|
5IGQ
 
 | |
5ID9
 
 | | Crystal structure of equine serum albumin in complex with phosphorodithioate derivative of myristoyl cyclic phosphatidic acid (cPA) | | Descriptor: | (4S)-2-sulfanylidene-4-[(tetradecanoyloxy)methyl]-1,3,2lambda~5~-dioxaphospholane-2-thiolate, FORMIC ACID, MALONATE ION, ... | | Authors: | Sekula, B, Bujacz, A, Rytczak, P, Bujacz, G. | | Deposit date: | 2016-02-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural evidence of the species-dependent albumin binding of the modified cyclic phosphatidic acid with cytotoxic properties.
Biosci.Rep., 36, 2016
|
|
1IWK
 
 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Mutant(112K) Cytochrome P450cam | | Descriptor: | CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
5IJN
 
 | | Composite structure of the inner ring of the human nuclear pore complex (32 copies of Nup205) | | Descriptor: | NUCLEAR PORE COMPLEX PROTEIN NUP155, NUCLEAR PORE COMPLEX PROTEIN NUP205, NUCLEAR PORE COMPLEX PROTEIN NUP54, ... | | Authors: | Kosinski, J, Mosalaganti, S, von Appen, A, Beck, M. | | Deposit date: | 2016-03-02 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21.4 Å) | | Cite: | Molecular architecture of the inner ring scaffold of the human nuclear pore complex.
Science, 352, 2016
|
|
1TGX
 
 | | X-RAY STRUCTURE AT 1.55 A OF TOXIN GAMMA, A CARDIOTOXIN FROM NAJA NIGRICOLLIS VENOM. CRYSTAL PACKING REVEALS A MODEL FOR INSERTION INTO MEMBRANES | | Descriptor: | CHLORIDE ION, GAMMA-CARDIOTOXIN | | Authors: | Bilwes, A, Rees, B, Moras, D. | | Deposit date: | 1993-11-24 | | Release date: | 1994-04-30 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure at 1.55 A of toxin gamma, a cardiotoxin from Naja nigricollis venom. Crystal packing reveals a model for insertion into membranes.
J.Mol.Biol., 239, 1994
|
|
5I6D
 
 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 5 [3-(3-(p-Tolyl)ureido) benzoic acid] | | Descriptor: | 3-{[(4-methylphenyl)carbamoyl]amino}benzoic acid, GLYCEROL, O-phosphoserine sulfhydrylase, ... | | Authors: | Schnell, R, Maric, S, Schneider, G. | | Deposit date: | 2016-02-16 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
1IH0
 
 | | Structure of the C-domain of Human Cardiac Troponin C in Complex with Ca2+ Sensitizer EMD 57033 | | Descriptor: | 5-[1-(3,4-DIMETHOXY-BENZOYL)-1,2,3,4-TETRAHYDRO-QUINOLIN-6-YL]-6-METHYL-3,6-DIHYDRO-[1,3,4]THIADIAZIN-2-ONE, CALCIUM ION, TROPONIN C, ... | | Authors: | Wang, X, Li, M.X, Spyracopoulos, L, Beier, N, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-domain of human cardiac troponin C in complex with the Ca2+ sensitizing drug EMD 57033.
J.Biol.Chem., 276, 2001
|
|
1SVC
 
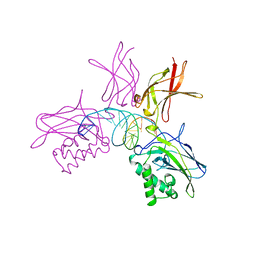 | | NFKB P50 HOMODIMER BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*CP*TP*A P*GP*A)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B (NF-KB)) | | Authors: | Mueller, C.W, Harrison, S.C. | | Deposit date: | 1995-11-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the NF-kappa B p50 homodimer bound to DNA.
Nature, 373, 1995
|
|
5I7A
 
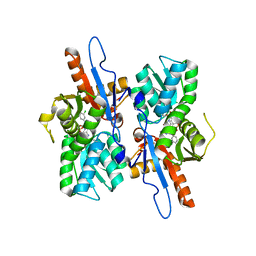 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 1 [3-(3-(3,4-Dichlorophenyl)ureido)benzoic acid] | | Descriptor: | 3-{[(3,4-dichlorophenyl)carbamoyl]amino}benzoic acid, O-phosphoserine sulfhydrylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schnell, R, Maric, S, Schneider, G. | | Deposit date: | 2016-02-17 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
1TLL
 
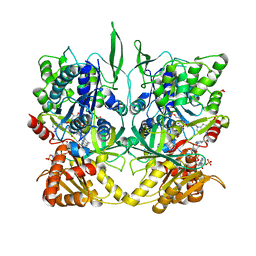 | | CRYSTAL STRUCTURE OF RAT NEURONAL NITRIC-OXIDE SYNTHASE REDUCTASE MODULE AT 2.3 A RESOLUTION. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Garcin, E.D, Bruns, C.M, Lloyd, S.J, Hosfield, D.J, Tiso, M, Gachhui, R, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase
J.Biol.Chem., 279, 2004
|
|
