9L1X
 
 | | hDEK-nucleosome complex (conformation 1) | | Descriptor: | 601 DNA (189-MER), 601 DNA_R (189-MER), Histone H2A type 1-B/E, ... | | Authors: | Liu, Y, Wang, C, Huang, H. | | Deposit date: | 2024-12-16 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | DEK-nucleosome structure shows DEK modulates H3K27me3 and stem cell fate.
Nat.Struct.Mol.Biol., 2025
|
|
9R04
 
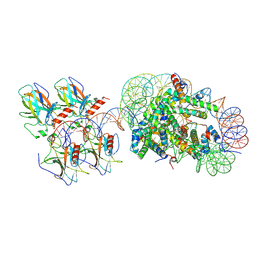 | | p53 bound to the nucleosome at position SHL-5.7 (crosslinked sample) | | Descriptor: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | Authors: | Chakraborty, D, Michael, A.K, Kempf, G, Cavadini, S, Kater, L, Thoma, N.H. | | Deposit date: | 2025-04-24 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | p53 bound to the nucleosome at position SHL-5.7 (crosslinked sample)
To Be Published
|
|
9R2M
 
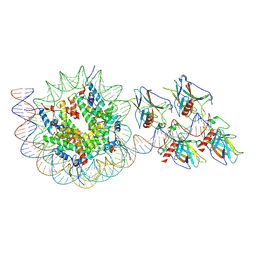 | | p53 bound to nucleosome at position SHL+5.9 (non-crosslinked sample, composite map) | | Descriptor: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | Authors: | Chakraborty, D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2025-04-30 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | p53 bound to nucleosome at position SHL+5.9 (non-crosslinked sample, composite map)
To Be Published
|
|
9N6I
 
 | | 2.61 A S.cerevisiae Chd1[L886G/L889G/L891G]-nucleosome 2:1 complex | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Folkwein, H.J, Armache, J.-P, Bowman, G.D. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | A competitive regulatory mechanism of the Chd1 remodeler is integral to distorting nucleosomal DNA.
Nat.Struct.Mol.Biol., 2025
|
|
4JJN
 
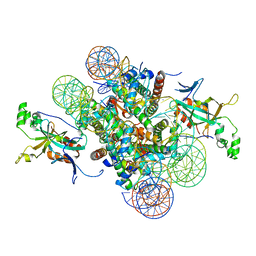 | | Crystal structure of heterochromatin protein Sir3 in complex with a silenced yeast nucleosome | | Descriptor: | DNA (146-MER), Histone H2A.2, Histone H2B.2, ... | | Authors: | Wang, F, Li, G, Mohammed, A, Lu, C, Currie, M, Johnson, A, Moazed, D. | | Deposit date: | 2013-03-08 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J8X
 
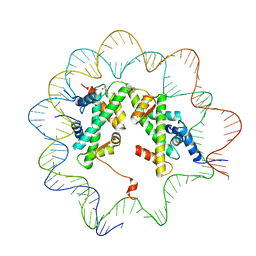 | |
4H9S
 
 | | Complex structure 6 of DAXX/H3.3(sub7)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4H9P
 
 | | Complex structure 3 of DAXX/H3.3(sub5,G90A)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4H9N
 
 | | Complex structure 1 of DAXX/H3.3(sub5)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4J8V
 
 | |
4J8U
 
 | |
4KGC
 
 | | Nucleosome Core Particle Containing (ETA6-P-CYMENE)-(1, 2-ETHYLENEDIAMINE)-RUTHENIUM | | Descriptor: | (ethane-1,2-diamine-kappa~2~N,N')[(1,2,3,4,5,6-eta)-1-methyl-4-(propan-2-yl)cyclohexane-1,2,3,4,5,6-hexayl]ruthenium, DNA (145-mer), Histone H2A, ... | | Authors: | Adhireksan, Z, Davey, C.A. | | Deposit date: | 2013-04-29 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Ligand substitutions between ruthenium-cymene compounds can control protein versus DNA targeting and anticancer activity
Nat Commun, 5, 2014
|
|
4KHA
 
 | | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Histone H2A, ... | | Authors: | Hondele, M, Halbach, F, Hassler, M, Ladurner, A.G. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT.
Nature, 499, 2013
|
|
4J8W
 
 | |
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8DU4
 
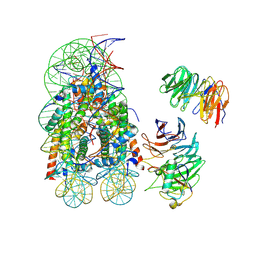 | | Complex between RbBP5-WDR5 and an H2B-ubiquitinated nucleosome | | Descriptor: | 601 DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Niklas, H.A, Rahman, S, Worden, E.J, Wolberger, C. | | Deposit date: | 2022-07-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Multistate structures of the MLL1-WRAD complex bound to H2B-ubiquitinated nucleosome.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8EUJ
 
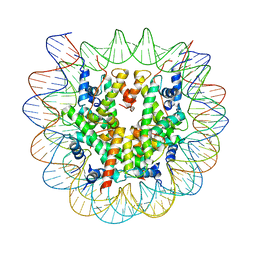 | | Class2 of the INO80-Nucleosome complex | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETT
 
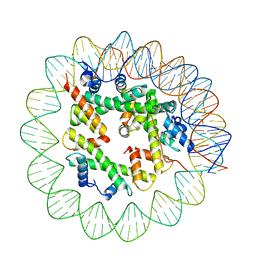 | | Class1 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (6.68 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETV
 
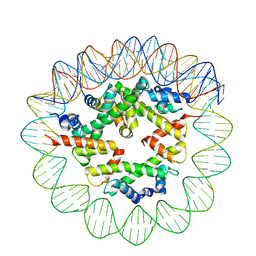 | | Class2 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EU2
 
 | | Class3 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EUE
 
 | | Class1 of the INO80-Nucleosome complex | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EVG
 
 | |
8EVJ
 
 | | CX3CR1 nucleosome bound PU.1 and C/EBPa | | Descriptor: | DNA (167-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Guan, R, Bai, Y, Lian, T. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBP alpha.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EVI
 
 | | CX3CR1 nucleosome and PU.1 complex containing disulfide bond mutations | | Descriptor: | DNA (167-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBP alpha.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EVH
 
 | | CX3CR1 nucleosome and wild type PU.1 complex | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBP alpha.
Nat.Struct.Mol.Biol., 31, 2024
|
|
