7ONM
 
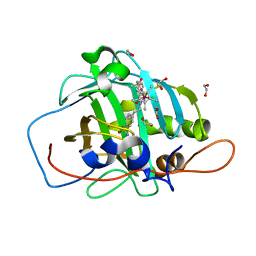 | | Carbonic anhydrase II mutant (N67G-E69R-I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
6TC1
 
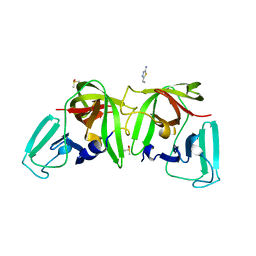 | |
9NA5
 
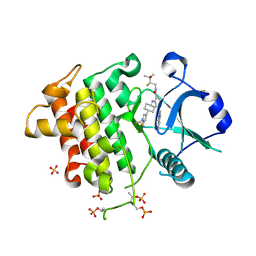 | | IRAK4 in Complex with Compound 24 | | Descriptor: | (6P)-6-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-4-({(1s,4S)-4-[1-(difluoromethyl)-1H-pyrazol-4-yl]cyclohexyl}amino)-N-[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]pyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 68, 2025
|
|
7ONQ
 
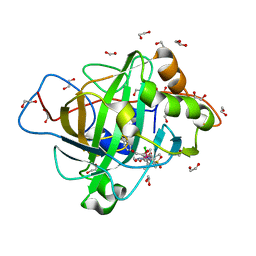 | | Carbonic anhydrase II mutant (E69C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONV
 
 | | Carbonic anhydrase II mutant (I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
9Q9S
 
 | | HSV-2 prefusion glycoprotein B bound by Nb1_gbHSV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, Nb1_gbHSV | | Authors: | Vollmer, B, Mulvaney, T, Ebel, H, Nentwig, J, Gruenewald, K. | | Deposit date: | 2025-02-26 | | Release date: | 2025-09-03 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A nanobody specific to prefusion glycoprotein B neutralizes HSV-1 and HSV-2.
Nature, 2025
|
|
8CN5
 
 | | Pa.FabF-C164Q in complex with propan-2-yl 1~{H}-pyrazole-3-carboxylate | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Georgiou, C, Brenk, R. | | Deposit date: | 2023-02-22 | | Release date: | 2024-01-10 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Towards new antibiotics: P. aeruginosa FabF ligands discovered by crystallographic fragment screening followed by hit expansion.
Eur.J.Med.Chem., 291, 2025
|
|
9O3B
 
 | | PKM2 bound to MCTI-566 | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, 7-[(dimethylamino)methyl]-8-fluoro-5-methyl-3-[(6-methylpyridin-2-yl)methyl]-3,5-dihydro-4H-pyridazino[4,5-b]indol-4-one, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2025-04-07 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | A Novel, Long-Acting, Small Molecule PKM2 Activator and Its Potential Broad Application Against Photoreceptor Degeneration.
Transl Vis Sci Technol, 14, 2025
|
|
9OA8
 
 | |
8UF5
 
 | | Catalytic domain of GtfB in complex with inhibitor G43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C, Velu, S. | | Deposit date: | 2023-10-03 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Discovery of Small Molecule Inhibitors of Cariogenic Virulence.
Sci Rep, 7, 2017
|
|
9R6R
 
 | |
6TGL
 
 | |
7M23
 
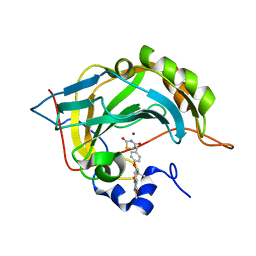 | | Human carbonic anhydrase II in complex with troglitazone | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, Carbonic anhydrase 2, ZINC ION | | Authors: | Mueller, S.L, Peat, T.S. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Glitazone Class of Drugs as Carbonic Anhydrase Inhibitors-A Spin-Off Discovery from Fragment Screening.
Molecules, 26, 2021
|
|
6TGV
 
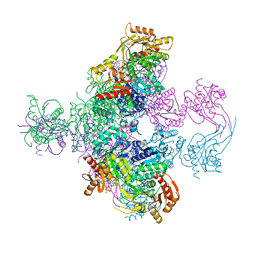 | | Crystal structure of Mycobacterium smegmatis CoaBC in complex with CTP and FMN | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-TRIPHOSPHATE, Coenzyme A biosynthesis bifunctional protein CoaBC, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
9R6Q
 
 | |
9NJF
 
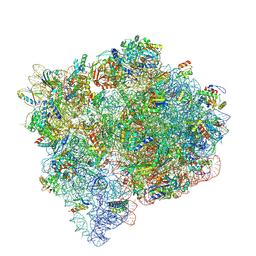 | |
7M26
 
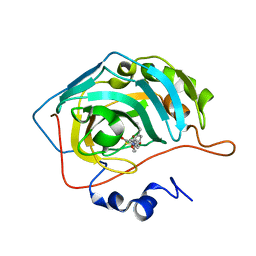 | | Human carbonic anhydrase II in complex with pioglitazone | | Descriptor: | (5R)-5-{4-[2-(5-ethylpyridin-2-yl)ethoxy]benzyl}-1,3-thiazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Mueller, S.L, Peat, T.S. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Glitazone Class of Drugs as Carbonic Anhydrase Inhibitors-A Spin-Off Discovery from Fragment Screening.
Molecules, 26, 2021
|
|
9R6P
 
 | |
9NTF
 
 | | Helix pomatia AMP deaminase (HPAMPD) with unknown density in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 301, 2025
|
|
6Y2O
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with 1,7-Naphthyridin-8-amine and PKI (5-24) | | Descriptor: | 1,7-naphthyridin-8-amine, DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Oebbeke, M, Heine, A, Klebe, G. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
9N3Q
 
 | |
9PCA
 
 | | HUMAN PRMT5:MEP50 COMPLEX IN COMPLEX WITH LIGAND 18 | | Descriptor: | (1S,2R,3S,5R)-3-{2-[2-amino-6-(2-hydroxyethyl)quinolin-7-yl]ethyl}-5-(7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclopentane-1,2-diol, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Milligan, C. | | Deposit date: | 2025-06-27 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Gut-Restricted PRMT5 Inhibitors to Intercept Colorectal Cancer in Patients with Genetic Loss of Tumor Suppressor Adenomatous Polyposis Coli.
J.Med.Chem., 2025
|
|
9R4O
 
 | | Crystal structure of inactive (D163N) variant CtGH76 from Chaetomium thermophilum in complex with alpha-1,6-mannobiose | | Descriptor: | GLYCEROL, Uncharacterized protein, alpha-D-mannopyranose, ... | | Authors: | Po-Hsun, W, Essen, L.-O. | | Deposit date: | 2025-05-07 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Ct GH76, a Glycoside Hydrolase 76 from Chaetomium thermophilum , with Elongated Glycan-Binding Canyon.
Int J Mol Sci, 26, 2025
|
|
9NTE
 
 | | Helix pomatia AMP deaminase (HPAMPD) apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 301, 2025
|
|
9ME1
 
 | |
