8PO4
 
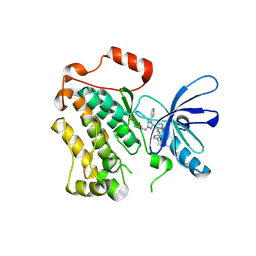 | |
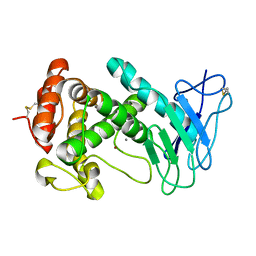
1EZM
 
 | |
8PNZ
 
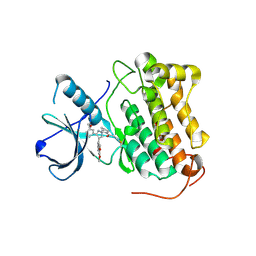 | |
5VH7
 
 | | Structure and dynamics of RNA repeat expansions that cause Huntington's Disease and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*CP*AP*GP*CP*AP*G)-3') | | Authors: | Chen, J.L, VanEtten, D.M, Fountain, M.A, Yildirim, I, Disney, M.D. | | Deposit date: | 2017-04-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of RNA Repeat Expansions That Cause Huntington's Disease and Myotonic Dystrophy Type 1.
Biochemistry, 56, 2017
|
|
7Q9L
 
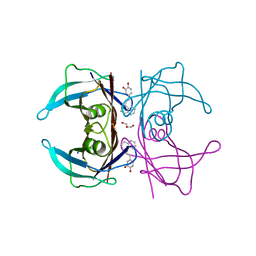 | | Transthyretin complexed with (E)-4-(2-(naphthalen-1-yl)vinyl)benzene-1,2-diol | | Descriptor: | 4-[(~{E})-2-naphthalen-1-ylethenyl]benzene-1,2-diol, GLYCEROL, SODIUM ION, ... | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
8EKG
 
 | | MHETase variant Thr159Val, Met192Tyr, Tyr252Phe, Tyr503Trp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saunders, J.W, Frkic, R.L, Jackson, C.J. | | Deposit date: | 2022-09-20 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Increasing the Soluble Expression and Whole-Cell Activity of the Plastic-Degrading Enzyme MHETase through Consensus Design.
Biochemistry, 63, 2024
|
|
6ORF
 
 | | Crystal structure of SpGH29 | | Descriptor: | 1,2-ETHANEDIOL, SpGH29, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-04-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two complementary alpha-fucosidases fromStreptococcus pneumoniaepromote complete degradation of host-derived carbohydrate antigens.
J.Biol.Chem., 294, 2019
|
|
5VH8
 
 | | Structure and dynamics of RNA repeat expansions that cause Huntington's Disease and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*CP*UP*GP*CP*UP*GP*CP*UP*GP*GP*UP*C)-3') | | Authors: | Chen, J.L, VanEtten, D.M, Fountain, M.A, Yildirim, I, Disney, M.D. | | Deposit date: | 2017-04-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of RNA Repeat Expansions That Cause Huntington's Disease and Myotonic Dystrophy Type 1.
Biochemistry, 56, 2017
|
|
9BEZ
 
 | | MID domain of human Argo2 bound to RNA | | Descriptor: | Protein argonaute-2, [(3~{S},4~{R},5~{R})-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-4-oxidanyl-oxolan-3-yl] [oxidanyl(phosphonooxy)phosphoryl] hydrogen phosphate | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
6PIZ
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-1 (NR02-24) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopropyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, GLYCEROL, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
8VDN
 
 | | DNA Ligase 1 with nick dG:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase 1, Downstream Oligo, ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-16 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
1F3T
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE (ODC) COMPLEXED WITH PUTRESCINE, ODC'S REACTION PRODUCT. | | Descriptor: | 1,4-DIAMINOBUTANE, ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jackson, L.K, Brooks, H.B, Osterman, A.L, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Altering the reaction specificity of eukaryotic ornithine decarboxylase.
Biochemistry, 39, 2000
|
|
8ZVM
 
 | |
7N9H
 
 | |
9AUB
 
 | | PLP-dependent BesB-F231Y variant holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, BesB F231Y variant, GLYCEROL, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2024-02-28 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Terminal alkyne formation by a pyridoxal phosphate-dependent enzyme.
Nat.Chem.Biol., 2025
|
|
7NU2
 
 | | Crystal Structure of Neisseria gonorrhoeae LeuRS E169G mutant in Complex with the Reaction Intermediate Leu-AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Leucine--tRNA ligase, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2021-03-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Partitioning of the initial catalytic steps of leucyl-tRNA synthetase is driven by an active site peptide-plane flip.
Commun Biol, 5, 2022
|
|
7NU4
 
 | | Crystal Structure of Neisseria gonorrhoeae LeuRS | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2021-03-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Partitioning of the initial catalytic steps of leucyl-tRNA synthetase is driven by an active site peptide-plane flip.
Commun Biol, 5, 2022
|
|
7NU8
 
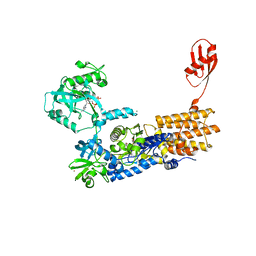 | | Crystal Structure of Neisseria gonorrhoeae LeuRS in Complex with the Reaction Intermediate Leu-AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Leucine--tRNA ligase, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2021-03-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Partitioning of the initial catalytic steps of leucyl-tRNA synthetase is driven by an active site peptide-plane flip.
Commun Biol, 5, 2022
|
|
7N91
 
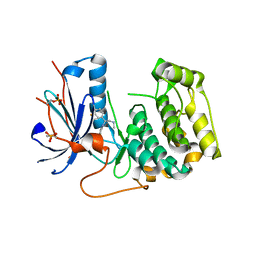 | | P70 S6K1 IN COMPLEX WITH MSC2317067A-1 | | Descriptor: | 4-{[(1S)-1-(3-fluorophenyl)-2-(methylamino)ethyl]amino}quinazoline-8-carboxamide, Ribosomal protein S6 kinase beta-1 | | Authors: | Mochalkin, I. | | Deposit date: | 2021-06-16 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of Clinical Candidate M2698, a Dual p70S6K and Akt Inhibitor, for Treatment of PAM Pathway-Altered Cancers.
J.Med.Chem., 64, 2021
|
|
7NU7
 
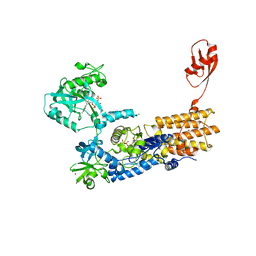 | | Crystal Structure of Neisseria gonorrhoeae LeuRS in Complex with ATP in Conformation 2 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Leucine--tRNA ligase, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2021-03-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Partitioning of the initial catalytic steps of leucyl-tRNA synthetase is driven by an active site peptide-plane flip.
Commun Biol, 5, 2022
|
|
7A22
 
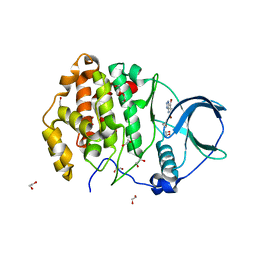 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7-tris(bromanyl)-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
4XMO
 
 | | Crystal structure of c-Met in complex with (R)-5-(8-fluoro-3-(1-fluoro-1-(3-methoxyquinolin-6-yl)ethyl)-[1,2,4]triazolo[4,3-a]pyridin-6-yl)-3-methylisoxazole | | Descriptor: | 6-{(1R)-1-fluoro-1-[8-fluoro-6-(3-methyl-1,2-oxazol-5-yl)[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}-3-methoxyquinoline, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Potent and Selective 8-Fluorotriazolopyridine c-Met Inhibitors.
J.Med.Chem., 58, 2015
|
|
8UHP
 
 | | anti-Phosphohistidine Fab hSC44.ck.20.N32F with 3pTZA peptide | | Descriptor: | 1,2-ETHANEDIOL, 3pTza peptide, ACETATE ION, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-09 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
8UIH
 
 | | anti-Phosphohistidine Fab hSC44.ck.20 with 3pTza peptide | | Descriptor: | 1,2-ETHANEDIOL, 3ptza peptide, hSC44.ck.20 Fab heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-10 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
8UHN
 
 | | anti-Phosphohistidine Fab hSC44.ck.20.N32F with 3pHis peptide | | Descriptor: | 1,2-ETHANEDIOL, 3pHis peptide, ACETATE ION, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-09 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
