6RWG
 
 | | Structure of HIV-1 CAcSP1NC mutant(W41A,M42A) interacting with maturation inhibitor EP39 | | Descriptor: | Gag polyprotein, ZINC ION | | Authors: | Chen, X, Coric, P, Larue, V, Nonin-Lecomte, S, Bouaziz, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The HIV-1 maturation inhibitor, EP39, interferes with the dynamic helix-coil equilibrium of the CA-SP1 junction of Gag.
Eur.J.Med.Chem., 204, 2020
|
|
6S0N
 
 | | A9 peptide derived from Herceptin fab binding region | | Descriptor: | GLN-ASP-VAL-ASN-THR-ALA-VAL-ALA-TRP | | Authors: | De Luca, S, Verdoliva, V, Saviano, M, Fattorusso, R, Diana, D. | | Deposit date: | 2019-06-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPR and NMR characterization of the molecular interaction between A9 peptide and a model system of HER2 receptor: A fragment approach for selecting peptide structures specific for their target.
J.Pept.Sci., 26, 2020
|
|
6SAI
 
 | |
5MPL
 
 | | hnRNP A1 RRM2 in complex with 5'-UCAGUU-3' RNA | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, RNA UCAGUU | | Authors: | Barraud, P, Allain, F.H.-T. | | Deposit date: | 2016-12-16 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7.
Elife, 6, 2017
|
|
6I3Z
 
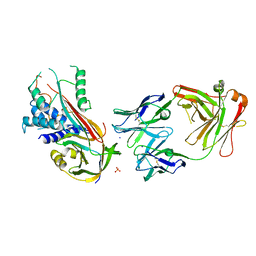 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin in complex with its antigen | | Descriptor: | Alpha-1-antitrypsin, Fab 2H2 heavy chain, Fab 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Jagger, A.M, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
3C13
 
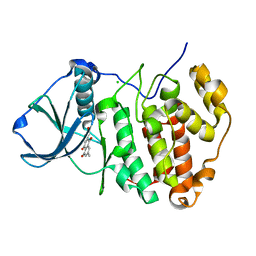 | | Low pH-value crystal structure of emodin in complex with the catalytic subunit of protein kinase CK2 | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Raaf, J, Issinger, O.-G. | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Catalytic Subunit of Human Protein Kinase CK2 Structurally Deviates from Its Maize Homologue in Complex with the Nucleotide Competitive Inhibitor Emodin
J.Mol.Biol., 377, 2008
|
|
7K59
 
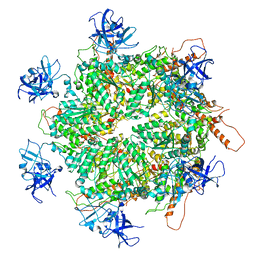 | | Structure of apo VCP hexamer generated from bacterially recombinant VCP/p97 | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6R9Z
 
 | |
6I7Z
 
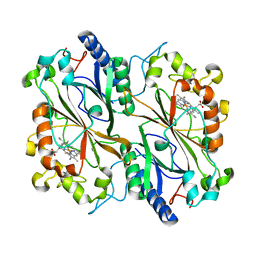 | | Dye type peroxidase Aa from Streptomyces lividans: 32.8 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-11-19 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
6I8E
 
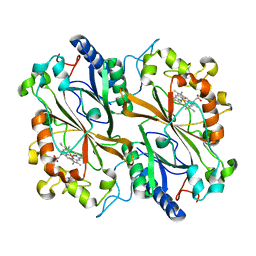 | | Dye type peroxidase Aa from Streptomyces lividans: 65.6 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-11-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
3C9A
 
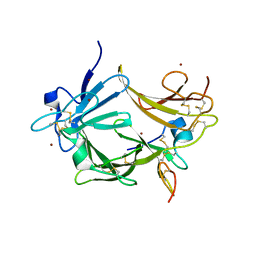 | | High Resolution Crystal Structure of Argos bound to the EGF domain of Spitz | | Descriptor: | BROMIDE ION, Protein giant-lens, Protein spitz | | Authors: | Klein, D.E, Stayrook, S.E, Shi, F, Narayan, K, Lemmon, M.A. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for EGFR ligand sequestration by Argos.
Nature, 453, 2008
|
|
8VLX
 
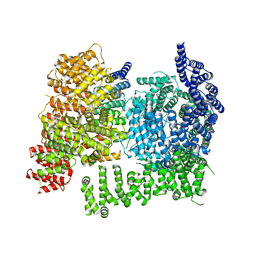 | | HTT in complex with HAP40 and a small molecule. | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin, N-[(1S,2R)-2-benzylcyclopentyl]-N'-{1-[(1S)-1-(pyridin-4-yl)ethyl]piperidin-4-yl}urea | | Authors: | Poweleit, N, Boudet, J, Doherty, E. | | Deposit date: | 2024-01-12 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Discovery of a Small Molecule Ligand to the Huntingtin/HAP40 complex
To Be Published
|
|
3CGU
 
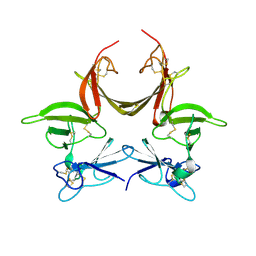 | |
6QYW
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - nisin ring A | | Descriptor: | ILE-DBU-DAL-ILE-DHA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2019-10-02 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6LHW
 
 | | Structure of N-terminal and C-terminal domains of FANCA | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.84 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
3CMZ
 
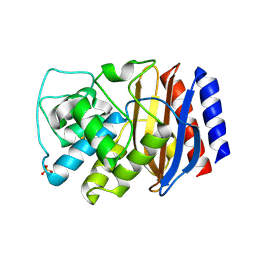 | | TEM-1 Class-A beta-lactamase L201P mutant apo structure | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Marciano, D.C, Wang, X, Wang, J, Chen, Y, Thomas, V.L, Shoichet, B.K, Palzkill, T. | | Deposit date: | 2008-03-24 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase
J.Mol.Biol., 384, 2008
|
|
8H05
 
 | |
8H03
 
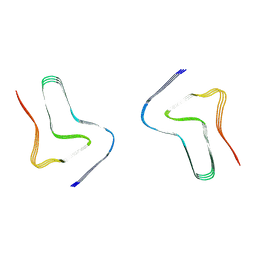 | |
8H04
 
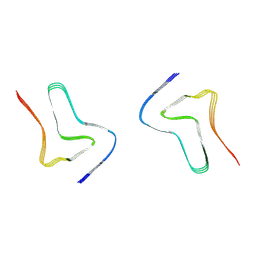 | |
6QFP
 
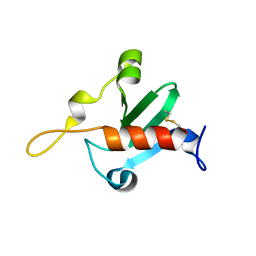 | |
6HNF
 
 | | Structure in solution of human fibronectin type III-domain 14 | | Descriptor: | Fibronectin | | Authors: | Zhong, X, Arnolds, O, Krenczyk, O, Gajewski, J, Puetz, S, Herrmann, C, Stoll, R. | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure in Solution of Fibronectin Type III Domain 14 Reveals Its Synergistic Heparin Binding Site.
Biochemistry, 57, 2018
|
|
6SKY
 
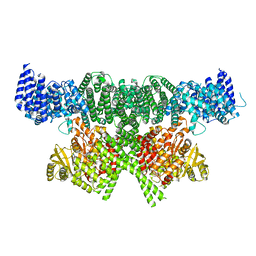 | | FAT and kinase domain of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
3AZJ
 
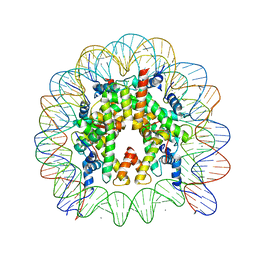 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K44Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
6SL0
 
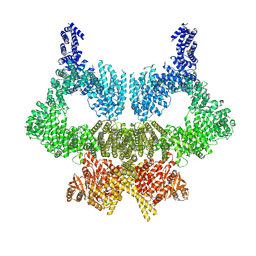 | | Complete CtTel1 dimer with C2 symmetry | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
