7MYA
 
 | |
5V86
 
 | | Structure of DCN1 bound to NAcM-OPT | | Descriptor: | Lysozyme,DCN1-like protein 1, N-benzyl-N-(1-butylpiperidin-4-yl)-N'-(3,4-dichlorophenyl)urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
7MYC
 
 | |
6U76
 
 | | Structure of methanesulfinate monooxygenase MsuC from Pseudomonas fluorescens. | | Descriptor: | methanesulfinate monooxygenase | | Authors: | Soule, J, Gnann, A.D, Parker, M.J, McKenna, K.C, Nguyen, S.V, Phan, N.T, Wicht, D.K, Dowling, D.P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | To be published
To Be Published
|
|
6HDH
 
 | |
5FOW
 
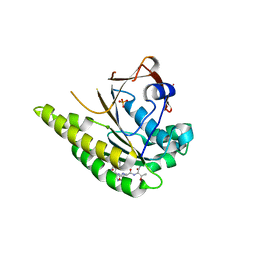 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH WHTA TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION, WHTA PEPTIDE | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
6RO5
 
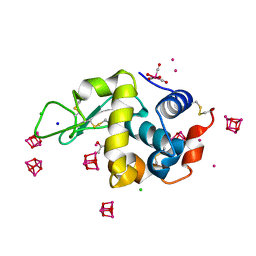 | | 1Yr-Y: Lysozyme with Re Cluster 1 year on shelf | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Brink, A, Helliwell, J.R. | | Deposit date: | 2019-05-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Formation of a highly dense tetra-rhenium cluster in a protein crystal and its implications in medical imaging.
Iucrj, 6, 2019
|
|
6ROK
 
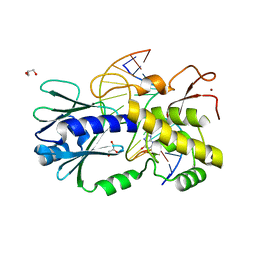 | | The crystal structure of a complex between the LlFpg protein, a THF-DNA and an inhibitor | | Descriptor: | 2,8-dithioxo-1,2,3,7,8,9-hexahydro-6H-purin-6-one, DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Coste, F, Goffinont, S, Castaing, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thiopurine Derivative-Induced Fpg/Nei DNA Glycosylase Inhibition: Structural, Dynamic and Functional Insights.
Int J Mol Sci, 21, 2020
|
|
3OGT
 
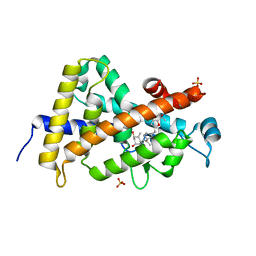 | | Design, Chemical synthesis, Functional characterization and Crystal structure of the sidechain analogue of 1,25-dihydroxyvitamin D3. | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-[5-(1-hydroxy-1-methylethyl)furan-2-yl]-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Huet, T, Fraga, R, Mourino, A, Moras, D, Rochel, N. | | Deposit date: | 2010-08-17 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, Chemical synthesis, Functional characterization and Crystal structure of the sidechain analogue of 1,25-dihydroxyvitamin D3.
To be Published
|
|
3NL7
 
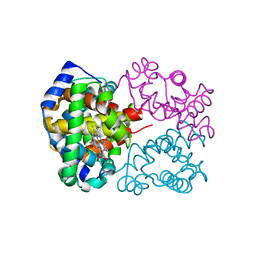 | | Human Hemoglobin A mutant beta H63W carbonmonoxy-form | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Birukou, I, Soman, J, Olson, J.S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Blocking the gate to ligand entry in human hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2V8G
 
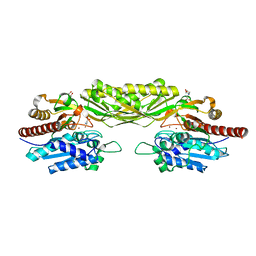 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri in complex with the product beta-alanine | | Descriptor: | BETA-ALANINE, BETA-ALANINE SYNTHASE, BICINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
6RIH
 
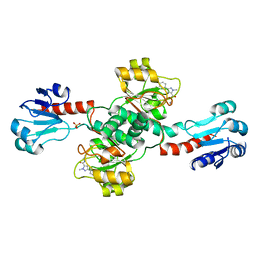 | | Crystal structure of PHGDH in complex with compound 9 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-cyclopropyl-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
5UL0
 
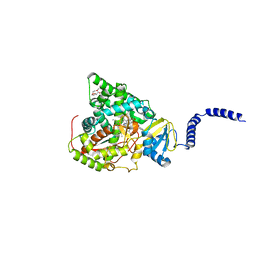 | | S. CEREVISIAE CYP51 COMPLEXED WITH VT-1161 | | Descriptor: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, Lanosterol 14-alpha demethylase, PENTAETHYLENE GLYCOL, ... | | Authors: | Tyndall, J.D, Keniya, M.V, Sabherwal, M, Monk, B.C. | | Deposit date: | 2017-01-23 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Azole Resistance Reduces Susceptibility to the Tetrazole Antifungal VT-1161.
Antimicrob.Agents Chemother., 63, 2019
|
|
8JU7
 
 | |
5UMH
 
 | |
2VBC
 
 | | Crystal structure of the NS3 protease-helicase from Dengue virus | | Descriptor: | DENGUE 4 NS3 FULL-LENGTH PROTEIN, PARTIAL POLYPROTEIN FOR NS2A AND NS2B, TYPE 4 PROTOTYPE DV4 H241 | | Authors: | Luo, D.H, Xu, T, Hunke, C, Gruber, G, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure of the Ns3 Protease-Helicase from Dengue Virus.
J.Virol., 82, 2008
|
|
5ULG
 
 | |
5FYG
 
 | | Structure of CYP153A from Marinobacter aquaeolei in complex with hydroxydodecanoic acid | | Descriptor: | 12-HYDROXYDODECANOIC ACID, CYTOCHROME P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Danesh-Azari, H.-R, Spandolf, C, Hoffman, S.M, Weissenborn, M, Hauer, B, Grogan, G. | | Deposit date: | 2016-03-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Guided Redesign of CYP153AM.aqfor the Improved Terminal Hydroxylation of Fatty Acids
Chemcatchem, 2016
|
|
2V7P
 
 | | Crystal structure of lactate dehydrogenase from Thermus Thermophilus HB8 (Holo form) | | Descriptor: | L-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID | | Authors: | Coquelle, N, Fioravanti, E, Weik, M, Vellieux, F. | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activity, stability and structural studies of lactate dehydrogenases adapted to extreme thermal environments.
J. Mol. Biol., 374, 2007
|
|
5FOV
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHTG TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, FHTG PEPTIDE, GLYCEROL, ... | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
6VCA
 
 | | TB38 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, ... | | Authors: | Zhou, Y.F, Lord, D.M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms.
J.Biol.Chem., 295, 2020
|
|
6RRX
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3R)-2-(Hydroxymethyl)-3-piperidinol | | Descriptor: | (2~{S},3~{R})-2-(hydroxymethyl)piperidin-3-ol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6VDE
 
 | | Full-length M. smegmatis Pol1 | | Descriptor: | DNA polymerase I, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
8JZJ
 
 | | E.coli Glyceraldehyde-3-phosphate dehydrogenase structure under cryoprotect condition of ammonium sulfate | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Jang, K, Hlaing, S.H.S, Kim, H.G, Kim, N, Choe, H.W, Kim, Y.J. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Strategy to Select an Appropriate Cryoprotectant for an X-ray Study of Escherichia coli GAPDH Crystals
Cryst.Growth Des., 23, 2023
|
|
6RJ2
 
 | | Crystal structure of PHGDH in complex with compound 40 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-[(1~{R})-1-[4-(ethanoylsulfamoyl)phenyl]ethyl]-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
