7F8C
 
 | |
5FJJ
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
7F8B
 
 | |
5FKS
 
 | | Unraveling the first step of xyloglucan degradation by the soil saprophyte Cellvibrio japonicus through the functional and structural characterization of a potent GH74 endo-xyloglucanase | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, ENDO-1,4-BETA-GLUCANASE/XYLOGLUCANASE, ... | | Authors: | Attia, M, Stepper, J, Davies, G.J, Brumer, H. | | Deposit date: | 2015-10-19 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Characterization of a Potent Gh74 Endo-Xyloglucanase from the Soil Saprophyte Cellvibrio Japonicus Unravels the First Step of Xyloglucan Degradation.
FEBS J., 283, 2016
|
|
7F2D
 
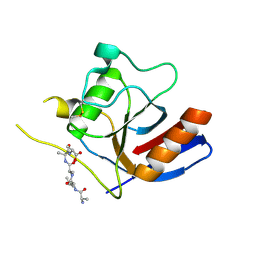 | |
7F2I
 
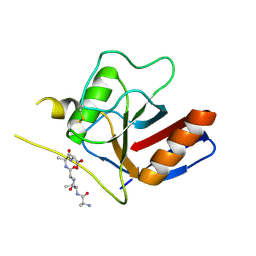 | |
5FHA
 
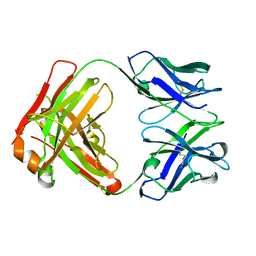 | |
5J96
 
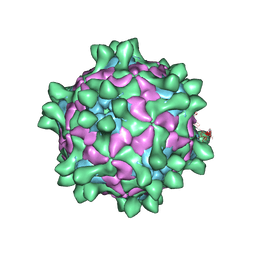 | | Crystal structure of Slow Bee Paralysis Virus at 3.4A resolution | | Descriptor: | Genome polyprotein, VP1, VP2 | | Authors: | Kalynych, S, Levdansky, Y, Palkova, L, Plevka, P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Virion Structure of Iflavirus Slow Bee Paralysis Virus at 2.6-Angstrom Resolution.
J.Virol., 90, 2016
|
|
5FHJ
 
 | |
7F8A
 
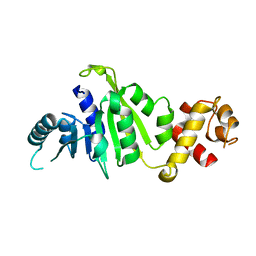 | | Crystal structure of rRNA methyltransferase Erm38 | | Descriptor: | Erm(38) | | Authors: | Goh, B.C, Lescar, J. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of mycobacterial erythromycin resistance methyltransferase Erm38 reveals its RNA-binding site.
J.Biol.Chem., 298, 2022
|
|
5WDE
 
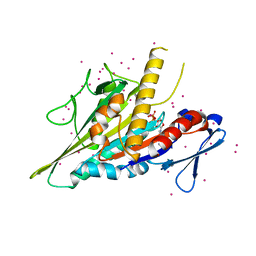 | | Crystal structure of the KIFC3 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIFC3, MAGNESIUM ION, ... | | Authors: | Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of small molecule ATPase inhibition of a human mitotic kinesin motor protein.
Sci Rep, 7, 2017
|
|
5FMB
 
 | |
7FFH
 
 | | Diarylpentanoid-producing polyketide synthase (N199L mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Takeshi, K, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
5FHV
 
 | | Crystal structure of mCherry after reaction with 2-mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, HEXAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2015-12-22 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Efficient switching of mCherry fluorescence using chemical caging.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7FFG
 
 | | Diarylpentanoid-producing polyketide synthase (N199F mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7FFA
 
 | | Diarylpentanoid-producing polyketide synthase from Aquilaria sinensis | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
5WMQ
 
 | |
5FRA
 
 | | CBM40_CPF0721-6'SL | | Descriptor: | ACETATE ION, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, ... | | Authors: | Ribeiro, J.P, Pau, W, Pifferi, C, Renaudet, O, Varrot, A, Mahal, L.K, Imberty, A. | | Deposit date: | 2015-12-16 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a High-Affinity Sialic Acid-Specific Cbm40 from Clostridium Perfringens and Engineering of a Divalent Form.
Biochem.J., 473, 2016
|
|
7FFI
 
 | | Diarylpentanoid-producing polyketide synthase (F340W mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
5WMW
 
 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | CYANIDE ION, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5ITI
 
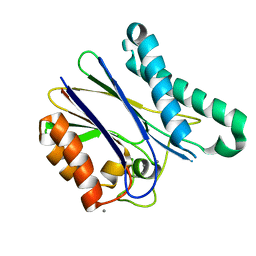 | | A cynobacterial PP2C (tPphA) structure | | Descriptor: | CALCIUM ION, Protein serin-threonin phosphatase | | Authors: | Su, J.Y. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Characterization of a Cyanobacterial PP2C Phosphatase Reveals Insights into Catalytic Mechanism and Substrate Recognition
Catalysts, 6, 2016
|
|
5FU2
 
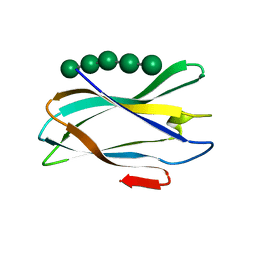 | | The complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition | | Descriptor: | CALCIUM ION, CBM74-RFGH5, SODIUM ION, ... | | Authors: | Basle, A, Luis, A.S, Venditto, I, Gilbert, H.J. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5WIK
 
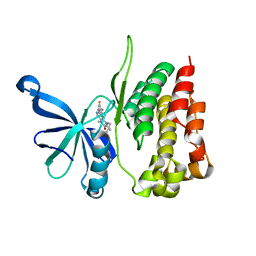 | | JAK2 Pseudokinase in complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIM
 
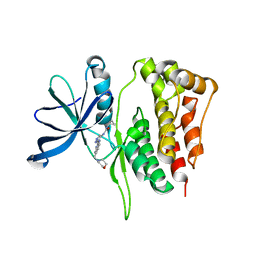 | | JAK2 Pseudokinase in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5FVC
 
 | | Structure of RNA-bound decameric HMPV nucleoprotein | | Descriptor: | HMPV NUCLEOPROTEIN, RNA | | Authors: | Renner, M, Bertinelli, M, Leyrat, C, Paesen, G.C, Saraiva de Oliveira, L.F, Huiskonen, J.T, Grimes, J.M. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein.
Elife, 5, 2016
|
|
