5VLA
 
 | | Short PCSK9 delta-P' complex with Fusion2 peptide | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-MET-PRO-TRP-ASN-LEU-VAL-ARG-ILE-GLY-LEU-LEU | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6WI1
 
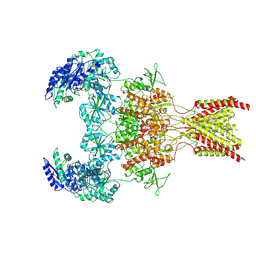 | | GluN1b-GluN2B NMDA receptor in active conformation stabilized by inter-GluN1b-GluN2B subunit cross-linking | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ionotropic glutamate receptor , ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
5VJ8
 
 | |
4OC7
 
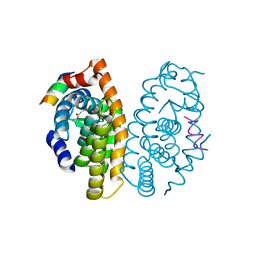 | | Retinoic acid receptor alpha in complex with (E)-3-(3'-allyl-6-hydroxy-[1,1'-biphenyl]-3-yl)acrylic acid and a fragment of the coactivator TIF2 | | Descriptor: | (2E)-3-[6-hydroxy-3'-(prop-2-en-1-yl)biphenyl-3-yl]prop-2-enoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Leysen, S, Scheepstra, M, Brunsveld, L, Milroy, L.G, Ottmann, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A natural-product switch for a dynamic protein interface.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6WKC
 
 | |
5VOX
 
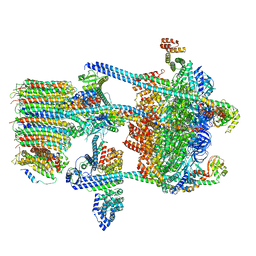 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 1) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
4ON9
 
 | | DECH box helicase domain | | Descriptor: | CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58, SULFATE ION | | Authors: | Deimling, T, Witte, G, Hopfner, K.P. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal and solution structure of the human RIG-I SF2 domain
Acta Crystallogr.,Sect.F, 70, 2014
|
|
6WC7
 
 | |
6WKW
 
 | |
6WL7
 
 | | Cryo-EM of Form 2 like peptide filament, 29-20-2 | | Descriptor: | peptide 29-20-2 | | Authors: | Wang, F, Gnewou, O.M, Modlin, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
5VE5
 
 | | Crystal structure of persulfide dioxygenase rhodanese fusion protein with rhodanese domain inactivating mutation (C314S) from Burkholderia phytofirmans in complex with glutathione | | Descriptor: | BpPRF, CHLORIDE ION, FE (III) ION, ... | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
6WDA
 
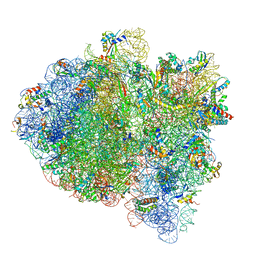 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure III-C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6WMT
 
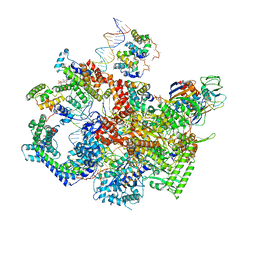 | | F. tularensis RNAPs70-(MglA-SspA)-ppGpp-PigR-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
5VFR
 
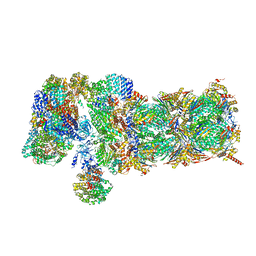 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VGM
 
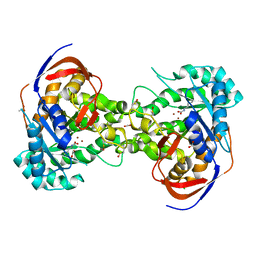 | | Crystal structure of dihydroorotase pyrC from Vibrio cholerae in complex with zinc at 1.95 A resolution. | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotase, ... | | Authors: | Lipowska, J, Shabalin, I.G, Miks, C.D, Winsor, J, Cooper, D.R, Shuvalova, L, Kwon, K, Lewinski, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-11 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
6WVR
 
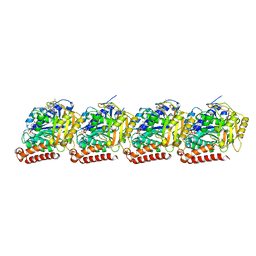 | | Tubulin dimers from a 13-protofilament, Taxol stabilized microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debs, G.E, Cha, M, Huehn, A.R, Sindelar, C.V. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Dynamic and asymmetric fluctuations in the microtubule wall captured by high-resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5VJM
 
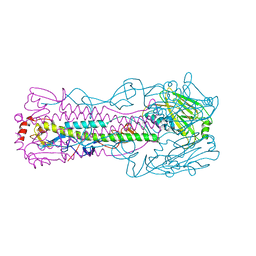 | | Crystal structure of H7 hemagglutinin mutant (V186K, K193T, G228S) from the influenza virus A/Shanghai/2/2013 (H7N9) with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2017-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Three mutations switch H7N9 influenza to human-type receptor specificity.
PLoS Pathog., 13, 2017
|
|
6WD8
 
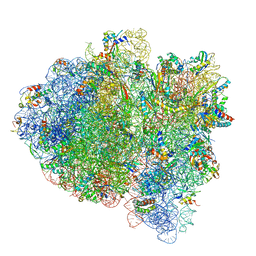 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure III-A) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
4O78
 
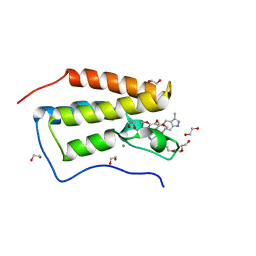 | | Crystal structure of the first bromodomain of human BRD4 in complex with GW612286X | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
6X3Y
 
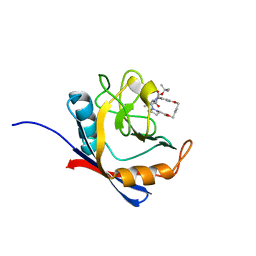 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, tert-butyl [(2S)-1-{[(3S,17S)-2,16-dioxo-10,15-dioxa-1,21-diazatricyclo[15.3.1.1~5,9~]docosa-5(22),6,8-trien-3-yl]amino}-3-methyl-1-oxobutan-2-yl]carbamate | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-21 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6WDO
 
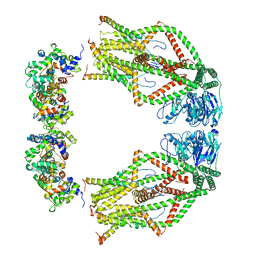 | | Cryo-EM structure of mitochondrial calcium uniporter holocomplex in high Ca2+ | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Feng, L, Zhang, J, Fan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and mechanism of the mitochondrial Ca2+uniporter holocomplex.
Nature, 582, 2020
|
|
4OK0
 
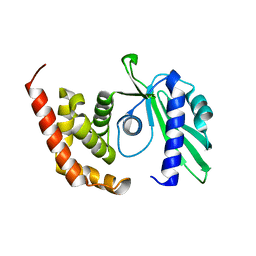 | | Crystal structure of putative nucleotidyltransferase from H. pylori | | Descriptor: | Putative | | Authors: | Yoon, J.Y, Lee, S.J, Lee, B, Yang, J.K, Suh, S.W. | | Deposit date: | 2014-01-21 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of JHP933 from Helicobacter pylori J99 shows two-domain architecture with a DUF1814 family nucleotidyltransferase domain and a helical bundle domain.
Proteins, 82, 2014
|
|
6X4N
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-3-oxa-9,12,15,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 24) | | Descriptor: | (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-3-oxa-9,12,15,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
5VKU
 
 | | An atomic structure of the human cytomegalovirus (HCMV) capsid with its securing layer of pp150 tegument protein | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tegument protein pp150, ... | | Authors: | Yu, X, Jih, J, Jiang, J, Zhou, H. | | Deposit date: | 2017-04-24 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150.
Science, 356, 2017
|
|
