5VLB
 
 | |
3P5D
 
 | | Structure of the carbohydrate-recognition domain of human Langerin with man5 (Man alpha1-3(Man alpha1-6)Man alpha1-6)(Man- alpha1-3)Man | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, alpha-D-mannopyranose, ... | | Authors: | Feinberg, H, Taylor, M.E, Razi, N, McBride, R, Knirel, Y.A, Graham, S.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8013 Å) | | Cite: | Structural Basis for Langerin Recognition of Diverse Pathogen and Mammalian Glycans through a Single Binding Site.
J.Mol.Biol., 405, 2011
|
|
1R5N
 
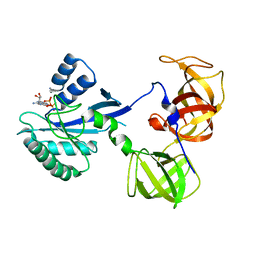 | | Crystal Structure Analysis of sup35 complexed with GDP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
1LUE
 
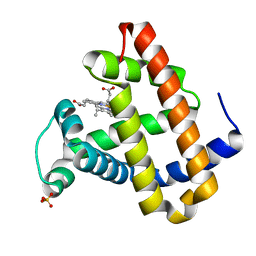 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68A/D122N MUTANT (MET) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-05-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
8IQI
 
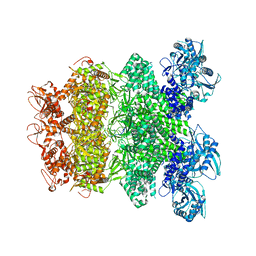 | | Structure of Full-Length AsfvPrimPol in Complex-Form | | Descriptor: | DNA (32-MER), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Shao, Z.W, Su, S.C, Gan, J.H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures and implications of the C962R protein of African swine fever virus.
Nucleic Acids Res., 51, 2023
|
|
8BFI
 
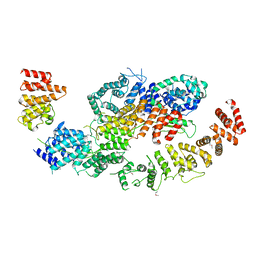 | | human CNOT1-CNOT10-CNOT11 module | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
5W7B
 
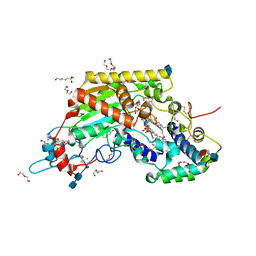 | | Rabbit acyloxyacyl hydrolase (AOAH), proteolytically processed, S262A mutant, with LPS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the mammalian lipopolysaccharide detoxifier.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3BSZ
 
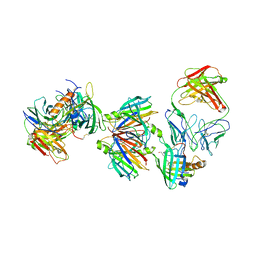 | | Crystal structure of the transthyretin-retinol binding protein-Fab complex | | Descriptor: | Fab fragment heavy chain, Fab fragment light chain, Plasma retinol-binding protein, ... | | Authors: | Zanotti, G, Cendron, L, Gliubich, F, Folli, C, Berni, R. | | Deposit date: | 2007-12-27 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
5C0P
 
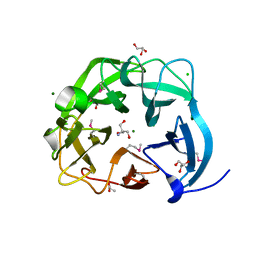 | | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endo-arabinase, ... | | Authors: | Tan, K, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482
To Be Published
|
|
6WKW
 
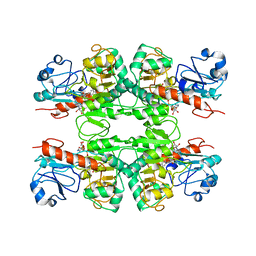 | |
3TS4
 
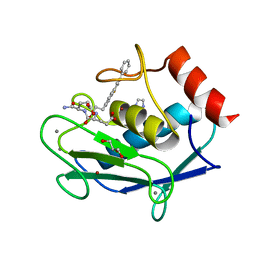 | | Human MMP12 in complex with L-glutamate motif inhibitor | | Descriptor: | CALCIUM ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Stura, E.A, Dive, V, Devel, L, Czarny, B, Beau, F, Vera, L. | | Deposit date: | 2011-09-12 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | Simple pseudo-dipeptides with a P2' glutamate: a novel inhibitor family of matrix metalloproteases and other metzincins.
J.Biol.Chem., 287, 2012
|
|
8IQH
 
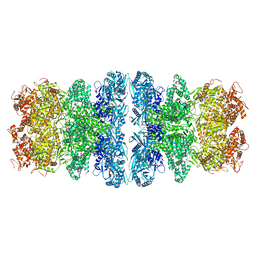 | |
8EAN
 
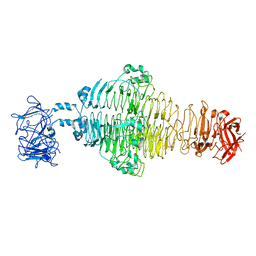 | |
5C3J
 
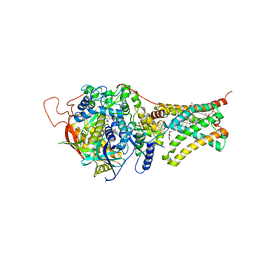 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with Ubiquinone-1 | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
To Be Published
|
|
1L2F
 
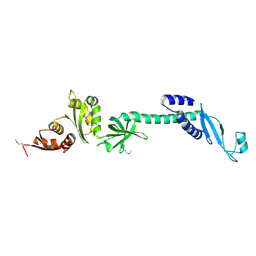 | | Crystal structure of NusA from Thermotoga maritima: a structure-based role of the N-terminal domain | | Descriptor: | N utilization substance protein A | | Authors: | Shin, D.H, Nguyen, H.H, Jancarik, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-02-20 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain.
Biochemistry, 42, 2003
|
|
3TLW
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' oxidized mutant in a locally-closed conformation (LC2 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1LE4
 
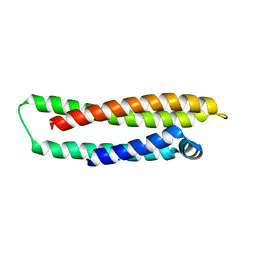 | |
1RDX
 
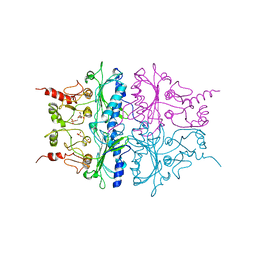 | | R-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
8B4O
 
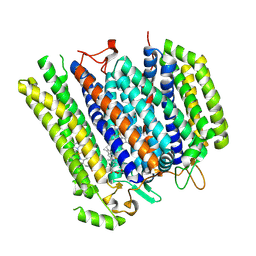 | | Cryo-EM structure of cytochrome bd oxidase from C. glutamicum | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome BD ubiquinol oxidase subunit I, Cytochrome bd-type quinol oxidase subunit II, ... | | Authors: | Grund, T.N, Kusumoto, T, Michel, H, Sakamoto, J, Safarian, S. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Cryo-EM structure of cytochrome bd oxidase from C. glutamicum
To be published
|
|
5IXF
 
 | | Solution structure of the STAM2 SH3 with AMSH derived peptide complex | | Descriptor: | STAM-binding protein, Signal transducing adapter molecule 2 | | Authors: | Hologne, M, Cantrelle, F.X, Trivelli, X, Walker, O. | | Deposit date: | 2016-03-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Reveals the Interplay among the AMSH SH3 Binding Motif, STAM2, and Lys63-Linked Diubiquitin.
J.Mol.Biol., 428, 2016
|
|
3TMA
 
 | | Crystal structure of TrmN from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, methyltransferase | | Authors: | Fislage, M, Roovers, M, Tuszynska, I, Bujnicki, J.M, Droogmans, L, Versees, W. | | Deposit date: | 2011-08-31 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life.
Nucleic Acids Res., 40, 2012
|
|
5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
8BFH
 
 | | CNOT11 | | Descriptor: | CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
3OUM
 
 | | Crystal Structure of toxoflavin-degrading enzyme in complex with toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, MANGANESE (II) ION, toxoflavin-degrading enzyme | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2010-09-15 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of phytotoxin toxoflavin-degrading enzyme
Plos One, 6, 2011
|
|
6WTI
 
 | | The Cryo-EM structure of the ubiquinol oxidase from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome o ubiquinol oxidase, ... | | Authors: | Su, C.-C. | | Deposit date: | 2020-05-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
