1VWN
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 4.8 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1S20
 
 | | A novel NAD binding protein revealed by the crystal structure of E. Coli 2,3-diketogulonate reductase (YiaK) NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82 | | Descriptor: | Hypothetical oxidoreductase yiaK, L(+)-TARTARIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-01-07 | | Release date: | 2004-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
1SCH
 
 | | PEANUT PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PEANUT PEROXIDASE, ... | | Authors: | Schuller, D.J, Poulos, T.L. | | Deposit date: | 1996-01-23 | | Release date: | 1996-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of peanut peroxidase.
Structure, 4, 1996
|
|
1VEF
 
 | | Acetylornithine aminotransferase from Thermus thermophilus HB8 | | Descriptor: | Acetylornithine/acetyl-lysine aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Three-Dimensional Strutcure of Acetylornithine aminotransferase from Thermus thermophilus HB8
To be Published
|
|
1VED
 
 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.9 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-30 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.9 angstroms resolution in space
To be Published
|
|
1BP1
 
 | | CRYSTAL STRUCTURE OF BPI, THE HUMAN BACTERICIDAL PERMEABILITY-INCREASING PROTEIN | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERICIDAL/PERMEABILITY-INCREASING PROTEIN | | Authors: | Beamer, L.J, Carroll, S.F, Eisenberg, D. | | Deposit date: | 1997-04-08 | | Release date: | 1997-09-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution.
Science, 276, 1997
|
|
1CUN
 
 | | CRYSTAL STRUCTURE OF REPEATS 16 AND 17 OF CHICKEN BRAIN ALPHA SPECTRIN | | Descriptor: | PROTEIN (ALPHA SPECTRIN) | | Authors: | Grum, V.L, Li, D, MacDonald, R.I, Mondragon, A. | | Deposit date: | 1999-08-20 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two repeats of spectrin suggest models of flexibility.
Cell(Cambridge,Mass.), 98, 1999
|
|
1VGM
 
 | |
1RZF
 
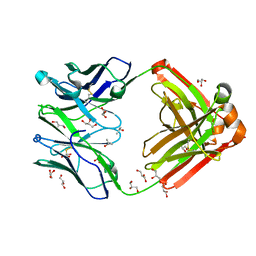 | | Crystal structure of Human anti-HIV-1 GP120-reactive antibody E51 | | Descriptor: | Fab E51 heavy chain, Fab E51 light chain, GLYCEROL, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2CBM
 
 | | Crystal structure of the apo-form of a neocarzinostatin mutant evolved to bind testosterone. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NEOCARZINOSTATIN | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
1VRK
 
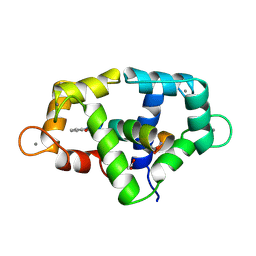 | |
1VJW
 
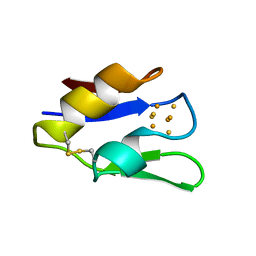 | | STRUCTURE OF OXIDOREDUCTASE (NADP+(A),FERREDOXIN(A)) | | Descriptor: | FERREDOXIN(A), IRON/SULFUR CLUSTER | | Authors: | Macedo-Ribeiro, S, Darimont, B, Sterner, R, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1996-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small structural changes account for the high thermostability of 1[4Fe-4S] ferredoxin from the hyperthermophilic bacterium Thermotoga maritima.
Structure, 4, 1996
|
|
2BVN
 
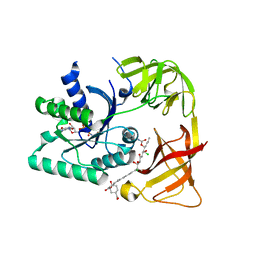 | | E. coli EF-Tu:GDPNP in complex with the antibiotic enacyloxin IIa | | Descriptor: | ELONGATION FACTOR TU, ENACYLOXIN IIA, MAGNESIUM ION, ... | | Authors: | Parmeggiani, A, Krab, I.M, Watanabe, T, Nielsen, R.C, Dahlberg, C, Nyborg, J, Nissen, P. | | Deposit date: | 2005-06-30 | | Release date: | 2005-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enacyloxin Iia Pinpoints a Binding Pocket of Elongation Factor TU for Development of Novel Antibiotics.
J.Biol.Chem., 281, 2006
|
|
1VOK
 
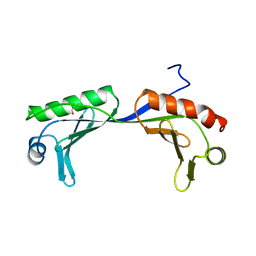 | | ARABIDOPSIS THALIANA TBP (DIMER) | | Descriptor: | SULFATE ION, TATA-BOX-BINDING PROTEIN | | Authors: | Nikolov, D.B, Burley, S.K. | | Deposit date: | 1996-04-29 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 A resolution refined structure of a TATA box-binding protein (TBP).
Nat.Struct.Biol., 1, 1994
|
|
1S5S
 
 | | Porcine trypsin complexed with guanidine-3-propanol inhibitor | | Descriptor: | CALCIUM ION, GLYCEROL, GUANIDINE-3-PROPANOL, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, DeRose, E.F, London, R.E. | | Deposit date: | 2004-01-21 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1VDC
 
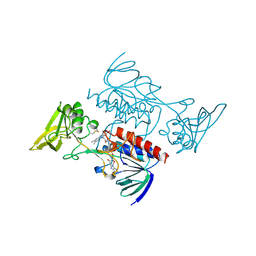 | | STRUCTURE OF NADPH DEPENDENT THIOREDOXIN REDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DEPENDENT THIOREDOXIN REDUCTASE, SULFATE ION | | Authors: | Dai, S, Eklund, H. | | Deposit date: | 1996-09-22 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Arabidopsis thaliana NADPH dependent thioredoxin reductase at 2.5 A resolution.
J.Mol.Biol., 264, 1996
|
|
1VDS
 
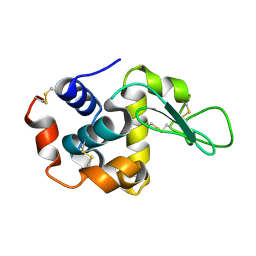 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space
to be published
|
|
1S70
 
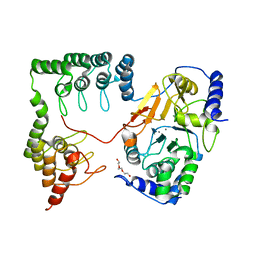 | | Complex between protein ser/thr phosphatase-1 (delta) and the myosin phosphatase targeting subunit 1 (MYPT1) | | Descriptor: | 130 kDa myosin-binding subunit of smooth muscle myosin phophatase (M130), MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-beta (or delta) catalytic subunit, ... | | Authors: | Kerff, F, Terrak, M, Dominguez, R. | | Deposit date: | 2004-01-28 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of protein phosphatase 1 regulation
Nature, 429, 2004
|
|
1VNH
 
 | |
1VFJ
 
 | | Crystal structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | nitrogen regulatory protein p-II | | Authors: | Wang, H, Sakai, H, Takemoto-Hori, C, Kaminishi, T, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8
J.STRUCT.BIOL., 149, 2005
|
|
2C3D
 
 | | 2.15 Angstrom crystal structure of 2-ketopropyl coenzyme M oxidoreductase carboxylase with a coenzyme M disulfide bound at the active site | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
1VFU
 
 | | Crystal structure of Thermoactinomyces vulgaris R-47 amylase 2/gamma-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
2BIF
 
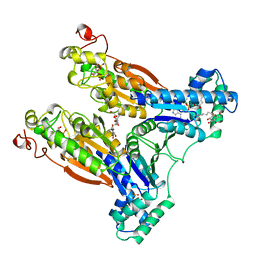 | | 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE H256A MUTANT WITH F6P IN PHOSPHATASE ACTIVE SITE | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Yuen, M.H, Hasemann, C.A. | | Deposit date: | 1998-10-26 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the H256A mutant of rat testis fructose-6-phosphate,2-kinase/fructose-2,6-bisphosphatase. Fructose 6-phosphate in the active site leads to mechanisms for both mutant and wild type bisphosphatase activities.
J.Biol.Chem., 274, 1999
|
|
2C4Y
 
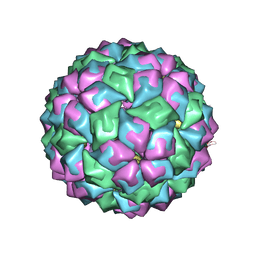 | | MS2-RNA HAIRPIN (2THIOURACIL-5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *SUR*AP*CP*CP*CP*AP*UP*GP*U)-3', Capsid protein | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-10-25 | | Release date: | 2006-01-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
2BKW
 
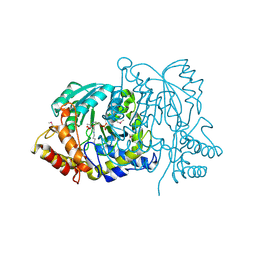 | | Yeast alanine:glyoxylate aminotransferase YFL030w | | Descriptor: | ALANINE-GLYOXYLATE AMINOTRANSFERASE 1, GLYOXYLIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meyer, P, Liger, D, Leulliot, N, Quevillon-Cheruel, S, Zhou, C.Z, Borel, F, Ferrer, J.L, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2005-02-21 | | Release date: | 2005-11-02 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure and Confirmation of the Alanine:Glyoxylate Aminotransferase Activity of the Yfl030W Yeast Protein
Biochimie, 87, 2005
|
|
