2XNA
 
 | | Crystal structure of the complex between human T cell receptor and staphylococcal enterotoxin | | Descriptor: | ENTEROTOXIN H, GLYCEROL, SODIUM ION, ... | | Authors: | Saline, M, Rodstrom, K.E.J, Fischer, G, Orekhov, V.Y, Karlsson, B.G, Lindkvist-Petersson, K. | | Deposit date: | 2010-07-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Superantigen Complexed with Tcr and Mhc Reveals Novel Insights Into Superantigenic T Cell Activation.
Nat.Commun., 1, 2010
|
|
1BMV
 
 | | PROTEIN-RNA INTERACTIONS IN AN ICOSAHEDRAL VIRUS AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (ICOSAHEDRAL VIRUS - A DOMAIN), PROTEIN (ICOSAHEDRAL VIRUS - B AND C DOMAIN), RNA (5'-R(*GP*GP*UP*CP*AP*AP*AP*AP*UP*GP*C)-3') | | Authors: | Chen, Z, Stauffacher, C, Li, Y, Schmidt, T, Bomu, W, Kamer, G, Shanks, M, Lomonossoff, G, Johnson, J.E. | | Deposit date: | 1989-10-09 | | Release date: | 1989-10-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein-RNA interactions in an icosahedral virus at 3.0 A resolution.
Science, 245, 1989
|
|
4YPC
 
 | |
4YAT
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-4-methoxybenzene-1-sulfonamide (5b) | | Descriptor: | N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-4-methoxybenzenesulfonamide, Transcription intermediary factor 1-alpha, ZINC ION | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YBT
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-{1,3-dimethyl-2-oxo-6-[3-(oxolan-3-ylmethoxy)phenoxy]-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1-methyl-1H-imidazole-4-sulfonamide (7l) | | Descriptor: | DIMETHYL SULFOXIDE, N-(1,3-dimethyl-2-oxo-6-{3-[(3S)-tetrahydrofuran-3-ylmethoxy]phenoxy}-2,3-dihydro-1H-benzimidazol-5-yl)-1-methyl-1H-imidazole-4-sulfonamide, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-19 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YC6
 
 | | CDK1/CKS1 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 1 | | Authors: | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2015-02-19 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
1BPE
 
 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA; EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA POLYMERASE BETA | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-04-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
7A29
 
 | |
7A5M
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-17]-OMe | | Descriptor: | Ac-[2-Cl-F]-[ProM-2]-[ProM-17]-OMe, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3JSS
 
 | |
4Y72
 
 | | Human CDK1/CyclinB1/CKS2 With Inhibitor | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
4A80
 
 | |
1HWI
 
 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH FLUVASTATIN | | Descriptor: | (3R,5S,6E)-7-[3-(4-fluorophenyl)-1-(propan-2-yl)-1H-indol-2-yl]-3,5-dihydroxyhept-6-enoic acid, ADENOSINE-5'-DIPHOSPHATE, HMG-COA REDUCTASE | | Authors: | Istvan, E.S, Deisenhofer, J. | | Deposit date: | 2001-01-09 | | Release date: | 2001-05-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mechanism for statin inhibition of HMG-CoA reductase.
Science, 292, 2001
|
|
1BPD
 
 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | DNA POLYMERASE BETA, PHOSPHATE ION | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-04-12 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
1BPB
 
 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-04-12 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
3B5U
 
 | | Actin filament model from extended form of acromsomal bundle in the Limulus sperm | | Descriptor: | Actin, alpha skeletal muscle | | Authors: | Cong, Y, Topf, M, Sali, A, Matsudaira, P, Dougherty, M, Chiu, W, Schmid, M.F. | | Deposit date: | 2007-10-26 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (9.5 Å) | | Cite: | Crystallographic conformers of actin in a biologically active bundle of filaments
J.Mol.Biol., 375, 2008
|
|
6I4T
 
 | | Crystal structure of the disease-causing I445M mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Wilk, P, Zambo, Z, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
6IV8
 
 | | the selenomethionine(SeMet)-derived Cas13d binary complex | | Descriptor: | MAGNESIUM ION, RNA (51-MER), RNA (53-MER), ... | | Authors: | Zhang, B, Ye, Y.M, Ye, W.W, OuYang, S.Y. | | Deposit date: | 2018-12-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d.
Nat Commun, 10, 2019
|
|
3O6M
 
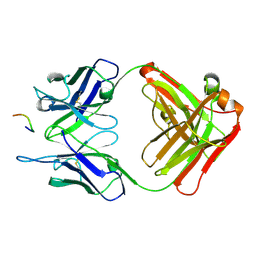 | | Anti-Tat HIV 11H6H1 Fab' complexed with a 9-mer Tat peptide | | Descriptor: | 11H6H1 Fab' heavy chain, 11H6H1 Fab' light chain, Protein Tat 9-mer peptide | | Authors: | Serriere, J, Gouet, P, Guillon, C. | | Deposit date: | 2010-07-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fab'-induced folding of antigenic N-terminal peptides from intrinsically disordered HIV-1 Tat revealed by X-ray crystallography.
J.Mol.Biol., 405, 2011
|
|
4WWA
 
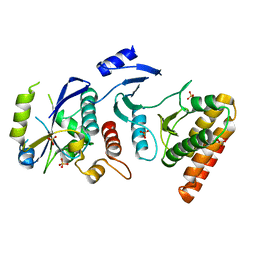 | | Crystal structure of binary complex Bud32-Cgi121 | | Descriptor: | EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, SULFATE ION | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4AG9
 
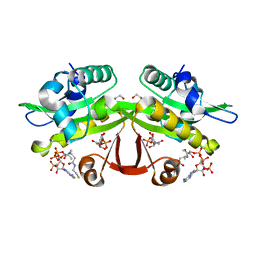 | | C. elegans glucosamine-6-phosphate N-acetyltransferase (GNA1): ternary complex with coenzyme A and GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, ... | | Authors: | Dorfmueller, H.C, Fang, W, Rao, F.V, Blair, D.E, Attrill, H, Shepherd, S.M, van Aalten, D.M.F. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Biochemical Characterization of a Trapped Coenzyme a Adduct of Caenorhabditis Elegans Glucosamine-6-Phosphate N-Acetyltransferase 1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1VND
 
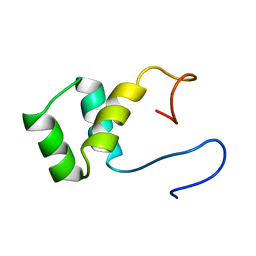 | | VND/NK-2 PROTEIN (HOMEODOMAIN), NMR | | Descriptor: | VND/NK-2 PROTEIN | | Authors: | Tsao, D.H.H, Gruschus, J.M, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1996-05-22 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the NK-2 homeodomain from Drosophila.
J.Mol.Biol., 251, 1995
|
|
3KTV
 
 | | Crystal structure of the human SRP19/S-domain SRP RNA complex | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2H64
 
 | |
4X6F
 
 | | CD1a binary complex with sphingomyelin | | Descriptor: | (4S,7S,23Z)-4-hydroxy-7-[(1S,2Z)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-9-oxo-3,5-dioxa-8-aza-4-phosphadotriacont- 23-en-1-aminium 4-oxide, Beta-2-microglobulin, T-cell surface glycoprotein CD1a | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
