6JTA
 
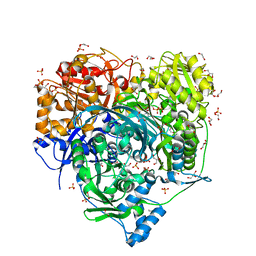 | | Crystal Structure of D464A L465A mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
1GUG
 
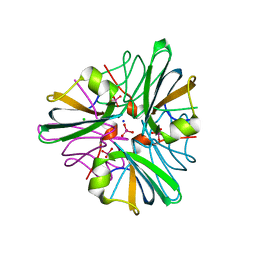 | | MopII from Clostridium pasteurianum complexed with tungstate | | Descriptor: | CHLORIDE ION, MOLYBDATE BINDING PROTEIN II, SODIUM ION, ... | | Authors: | Schuettelkopf, A.W, Harrison, J.A, Hunter, W.N. | | Deposit date: | 2002-01-25 | | Release date: | 2002-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Passive Acquisition of Ligand by the Mopii Molbindin from Clostridium Pasteurianum: Structures of Apo and Oxyanion-Bound Forms
J.Biol.Chem., 277, 2002
|
|
5JX4
 
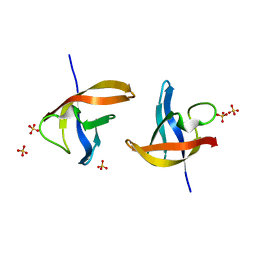 | | Crystal structure of E36-G37del mutant of the Bacillus caldolyticus cold shock protein. | | Descriptor: | Cold shock protein CspB, SULFATE ION | | Authors: | Carvajal, A, Castro-Fernandez, V, Cabrejos, D, Fuentealba, M, Pereira, H.M, Vallejos, G, Cabrera, R, Garratt, R.C, Komives, E.A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual dimerization of a BcCsp mutant leads to reduced conformational dynamics.
FEBS J., 284, 2017
|
|
5JTB
 
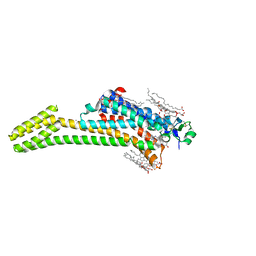 | | Crystal structure of the chimeric protein of A2aAR-BRIL with bound iodide ions | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Melnikov, I, Polovinkin, V, Shevtsov, M, Borshchevskiy, V, Cherezov, V, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
1UYS
 
 | |
6X28
 
 | | Crystal structure of PT2243 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | (1R)-4-(3,5-difluorophenoxy)-7-(trifluoromethyl)-2,3-dihydro-1H-inden-1-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of PT2243 bound to HIF2-PasB*:ARNT-PasB* complex
To Be Published
|
|
6KEV
 
 | | Reduced phosphoribulokinase from Synechococcus elongatus PCC 7942 complexed with adenosine diphosphate and glucose 6-phosphate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yu, A, Xie, Y, Li, M. | | Deposit date: | 2019-07-05 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.506139 Å) | | Cite: | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
5L1E
 
 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with noncompetitive inhibitor CP465022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-2-(2-{6-[(diethylamino)methyl]pyridin-2-yl}ethyl)-6-fluoroquinazolin-4(3H)-one, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.37 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
6X37
 
 | | Crystal structure of PT3245 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-fluoro-5-{[(7R)-7-hydroxy-1-(trifluoromethyl)-6,7-dihydro-5H-cyclopenta[c]pyridin-4-yl]oxy}benzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-21 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of PT3245 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
6X1J
 
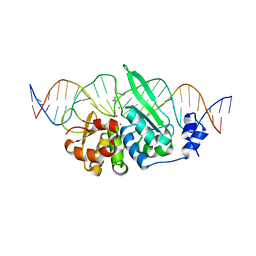 | |
6CGL
 
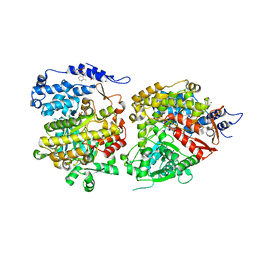 | |
6X3D
 
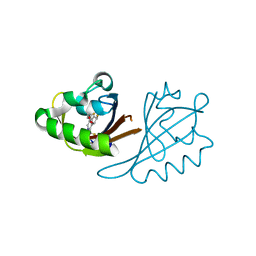 | | Crystal structure of PT3388 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | (6R,7S)-4-[(3,3-difluorocyclobutyl)oxy]-6-fluoro-1-(trifluoromethyl)-6,7-dihydro-5H-cyclopenta[c]pyridin-7-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-21 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PT3388 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
6O9M
 
 | | Structure of the human apo TFIIH | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription factor IIH subunit 1, General transcription factor IIH subunit 2, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7AOA
 
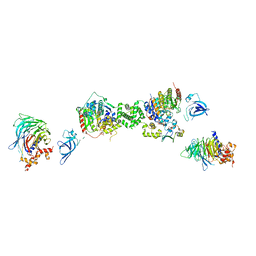 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
6X21
 
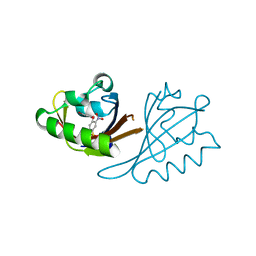 | | Crystal structure of PT1673 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 1-(3-bromo-5-fluorophenoxy)-4-[(difluoromethyl)sulfonyl]-2-nitrobenzene, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-19 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of PT1673 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
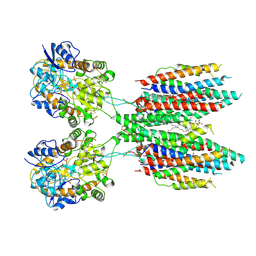
6UD8
 
 | |
7AUD
 
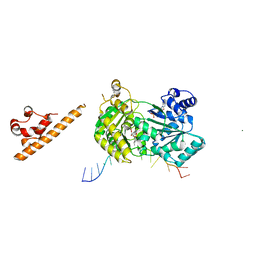 | |
7KQ5
 
 | |
1ZV1
 
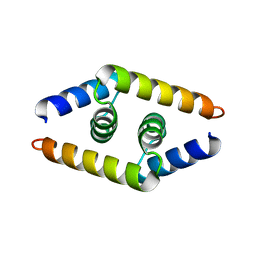 | | Crystal structure of the dimerization domain of doublesex protein from D. melanogaster | | Descriptor: | Doublesex protein | | Authors: | Weiss, M.A, Bayrer, J.R, Wan, Z, Li, B, Phillips, N.B. | | Deposit date: | 2005-06-01 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dimerization of doublesex is mediated by a cryptic ubiquitin-associated domain fold: implications for sex-specific gene regulation
J.Biol.Chem., 280, 2005
|
|
5LF5
 
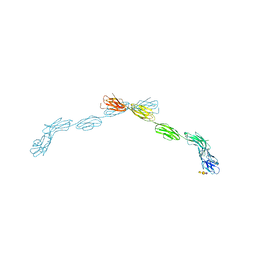 | |
6KEZ
 
 | | Crystal structure of GAPDH/CP12/PRK complex from Arabidopsis thaliana | | Descriptor: | Calvin cycle protein CP12-2, Glyceraldehyde-3-phosphate dehydrogenase GAPA1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yu, A, Xie, Y, Li, M. | | Deposit date: | 2019-07-05 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
6CQM
 
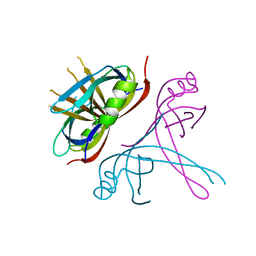 | | Crystal Structure of mitochondrial single-stranded DNA binding proteins from S. cerevisiae, Rim1 (Form2) | | Descriptor: | Single-stranded DNA-binding protein RIM1, mitochondrial | | Authors: | Singh, S.P, Kukshal, V, Bona, P.D, Lytle, A.K, Edwin, A, Galletto, R. | | Deposit date: | 2018-03-15 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mitochondrial single-stranded DNA binding protein from S. cerevisiae, Rim1, does not form stable homo-tetramers and binds DNA as a dimer of dimers.
Nucleic Acids Res., 46, 2018
|
|
2KNC
 
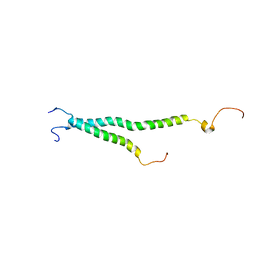 | | Platelet integrin ALFAIIB-BETA3 transmembrane-cytoplasmic heterocomplex | | Descriptor: | Integrin alpha-IIb, Integrin beta-3 | | Authors: | Yang, J, Ma, Y, Page, R.C, Misra, S, Plow, E.F, Qin, J. | | Deposit date: | 2009-08-20 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6OG3
 
 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, NTD-trimer | | Descriptor: | Alpha S1-casein, Hyperactive disaggregase ClpB | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OAY
 
 | | Structure of the hyperactive ClpB mutant K476C, bound to casein, post-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Hyperactive disaggregase ClpB, ... | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
