7SP7
 
 | | Chlorella virus hyaluronan synthase inhibited by UDP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7JSQ
 
 | | Refined structure of the C-terminal domain of DNAJB6b | | Descriptor: | DnaJ homolog subfamily B member 6 | | Authors: | Karamanos, T.K, Clore, G.M. | | Deposit date: | 2020-08-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An S/T motif controls reversible oligomerization of the Hsp40 chaperone DNAJB6b through subtle reorganization of a beta sheet backbone.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7T01
 
 | | SARS-CoV-2 S-RBD + Fab 54042-4 | | Descriptor: | 54042-4 Fab - Heavy Chain, 54042-4 Fab - Light Chain, Spike protein S1 | | Authors: | Johnson, N.V, Mclellan, J.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Potent neutralization of SARS-CoV-2 variants of concern by an antibody with an uncommon genetic signature and structural mode of spike recognition.
Cell Rep, 37, 2021
|
|
7TR8
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7SXK
 
 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - Full Length | | Descriptor: | Portal protein | | Authors: | Hou, C.F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-23 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
7TR9
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
5FR3
 
 | | X-ray crystal structure of aggregation-resistant protective antigen of Bacillus anthracis (mutant S559L T576E) | | Descriptor: | CALCIUM ION, GLYCEROL, PROTECTIVE ANTIGEN | | Authors: | Ganesan, A, Siekierska, A, Beerten, J, Brams, M, van Durme, J, De Baets, G, van der Kant, R, Gallardo, R, Ramakers, M, Langenberg, T, Wilkinson, H, De Smet, F, Ulens, C, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural Hot Spots for the Solubility of Globular Proteins
Nat.Commun., 7, 2016
|
|
6PVK
 
 | | Bacterial 45SRbgA ribosomal particle class A | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Ortega, J, Seffouh, A, Jain, N, Jahagirdar, D, Basu, K, Razi, A, Ni, X, Guarne, A, Britton, R.A. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate.
Nucleic Acids Res., 47, 2019
|
|
6BNO
 
 | | Structure of bare actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gurel, P.S, Alushin, G.A. | | Deposit date: | 2017-11-17 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity.
Elife, 6, 2017
|
|
6BNQ
 
 | |
5F81
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2015-12-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
7UWR
 
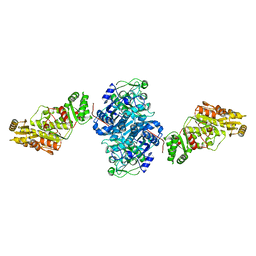 | | KSQ+AT from first module of the pikromycin synthase | | Descriptor: | Narbonolide/10-deoxymethynolide synthase PikA1, modules 1 and 2 | | Authors: | Keatinge-Clay, A.T, Dickinson, M.S, Miyazawa, T, McCool, R.S. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Priming enzymes from the pikromycin synthase reveal how assembly-line ketosynthases catalyze carbon-carbon chemistry.
Structure, 30, 2022
|
|
3TEZ
 
 | |
3TAB
 
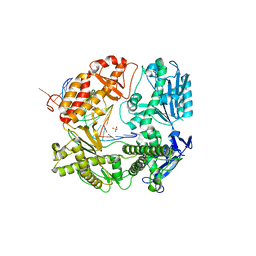 | | 5-hydroxycytosine paired with dGMP in RB69 gp43 | | Descriptor: | DNA (5'-D(*CP*CP*(5OC)P*GP*GP*TP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*AP*CP*CP*G)-3'), DNA polymerase, ... | | Authors: | Zahn, K.E. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The miscoding potential of 5-hydroxycytosine arises due to template instability in the replicative polymerase active site.
Biochemistry, 50, 2011
|
|
3TEY
 
 | |
5HL4
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | COBALT HEXAMMINE(III), FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
7U2B
 
 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(GCU-TL) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, RNA (53-MER), Serine--tRNA ligase, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
6CGR
 
 | |
7SEE
 
 | | Structure of E. coli LetB delta (Ring6) mutant, Ring1 in the closed state (Model 1) | | Descriptor: | MCE family protein, Intermembrane transport protein YebT chimera, UNKNOWN ATOM OR ION | | Authors: | Vieni, C, Coudray, N, Bhabha, G, Ekiert, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Role of Ring6 in the Function of the E. coli MCE Protein LetB.
J.Mol.Biol., 434, 2022
|
|
7SEF
 
 | | Structure of E. coli LetB delta (Ring6) mutant, Ring 1 in the open state (Model 2, Rings 1-3 only) | | Descriptor: | MCE family protein, Intermembrane transport protein YebT chimera | | Authors: | Vieni, C, Coudray, N, Bhabha, G, Ekiert, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Role of Ring6 in the Function of the E. coli MCE Protein LetB.
J.Mol.Biol., 434, 2022
|
|
7SQH
 
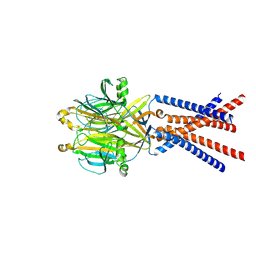 | |
7SQF
 
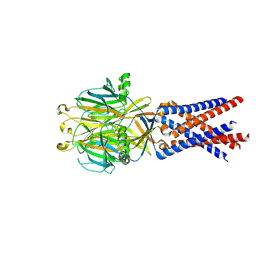 | |
7SQG
 
 | |
7ZKD
 
 | | The NMR structure of the MAX47 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-09-04 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7WAF
 
 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS and 4x(beta-Asp-Arg) | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, Cyanophycin synthase, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
